Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
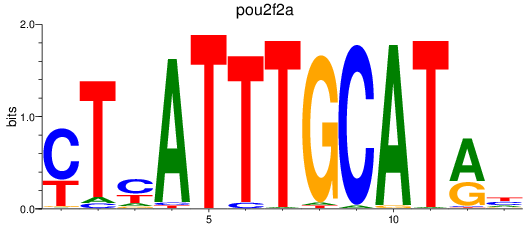
Results for pou2f2a
Z-value: 0.87
Transcription factors associated with pou2f2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f2a
|
ENSDARG00000019658 | POU class 2 homeobox 2a |
|
pou2f2a
|
ENSDARG00000036816 | POU class 2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f2a | dr11_v1_chr19_-_6239248_6239248 | -0.99 | 2.6e-14 | Click! |
Activity profile of pou2f2a motif
Sorted Z-values of pou2f2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_22320738 | 4.80 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr1_-_51038885 | 3.81 |
ENSDART00000035150
|
spast
|
spastin |
| chr5_-_65662996 | 3.38 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr17_-_4245311 | 2.80 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr23_+_7692042 | 2.77 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr17_-_4245902 | 2.32 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr4_-_9780931 | 2.16 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr22_+_980290 | 2.04 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr1_-_45320126 | 1.93 |
ENSDART00000133572
|
si:ch73-90k17.1
|
si:ch73-90k17.1 |
| chr24_-_25428176 | 1.92 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr5_+_26212621 | 1.88 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr21_+_7605803 | 1.86 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr14_-_46198373 | 1.85 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr9_+_2452672 | 1.82 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr24_-_31904924 | 1.77 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr4_-_4612116 | 1.72 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr17_-_6641535 | 1.67 |
ENSDART00000154540
ENSDART00000180384 |
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr14_-_45967981 | 1.66 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr17_-_6618574 | 1.62 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr3_-_15119856 | 1.62 |
ENSDART00000138328
|
xpo6
|
exportin 6 |
| chr14_-_16810401 | 1.50 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr1_-_55118745 | 1.48 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr9_+_54039006 | 1.37 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr14_+_35428152 | 1.35 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr7_-_26497947 | 1.32 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr5_+_16117871 | 1.30 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr3_-_32337653 | 1.28 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr15_-_931815 | 1.25 |
ENSDART00000106627
ENSDART00000102239 ENSDART00000184754 ENSDART00000186733 |
zgc:162936
|
zgc:162936 |
| chr8_-_2616326 | 1.23 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr6_+_4299164 | 1.23 |
ENSDART00000159759
|
nbeal1
|
neurobeachin-like 1 |
| chr3_+_7771420 | 1.23 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr9_-_27398369 | 1.20 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr18_+_22174630 | 1.18 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr9_+_19623363 | 1.18 |
ENSDART00000142471
ENSDART00000147662 ENSDART00000136053 |
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr17_+_23311377 | 1.17 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr10_+_33393829 | 1.17 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr9_+_22634073 | 1.14 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr21_-_2185004 | 1.13 |
ENSDART00000163405
|
zgc:171220
|
zgc:171220 |
| chr12_+_47794089 | 1.13 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr14_-_45967712 | 1.11 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr21_-_2185600 | 1.11 |
ENSDART00000169897
|
zgc:171220
|
zgc:171220 |
| chr18_-_3520358 | 1.10 |
ENSDART00000181412
|
capn5a
|
calpain 5a |
| chr5_-_63302944 | 1.10 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr3_-_27646070 | 1.09 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr16_+_4078240 | 1.08 |
ENSDART00000160890
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr16_-_2870522 | 1.06 |
ENSDART00000148543
|
cdcp1a
|
CUB domain containing protein 1a |
| chr13_+_38817871 | 1.03 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr10_+_3145707 | 1.00 |
ENSDART00000160046
|
hic2
|
hypermethylated in cancer 2 |
| chr23_-_12453700 | 1.00 |
ENSDART00000091140
|
snx21
|
sorting nexin family member 21 |
| chr6_-_59357256 | 1.00 |
ENSDART00000074534
|
fam210b
|
family with sequence similarity 210, member B |
| chr3_-_55525627 | 0.99 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr3_-_21118969 | 0.99 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr11_+_19433936 | 0.99 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr8_+_50190742 | 0.99 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr12_+_3571770 | 0.98 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr12_+_16087077 | 0.97 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr3_-_26190804 | 0.97 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr18_-_12957451 | 0.95 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr9_+_29431763 | 0.95 |
ENSDART00000186095
ENSDART00000182640 |
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr2_+_47718605 | 0.93 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr6_+_43450221 | 0.92 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr16_+_50434668 | 0.92 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr21_+_34122801 | 0.90 |
ENSDART00000182627
|
hmgb3b
|
high mobility group box 3b |
| chr19_+_32257472 | 0.88 |
ENSDART00000186471
|
atxn1a
|
ataxin 1a |
| chr5_+_33287611 | 0.88 |
ENSDART00000125093
ENSDART00000146759 |
med22
|
mediator complex subunit 22 |
| chr24_+_29912509 | 0.87 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr9_-_30502010 | 0.86 |
ENSDART00000149483
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr15_-_17138640 | 0.86 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr15_+_21262917 | 0.86 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr23_+_2825940 | 0.86 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr1_+_1712140 | 0.85 |
ENSDART00000081047
|
atp1a1a.1
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 1 |
| chr22_-_10397600 | 0.85 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr1_-_51720633 | 0.85 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr6_-_6258451 | 0.85 |
ENSDART00000081966
ENSDART00000125918 |
rtn4a
|
reticulon 4a |
| chr7_+_13824150 | 0.84 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr22_-_3299100 | 0.84 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr15_-_1622468 | 0.84 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr13_-_49444636 | 0.83 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr5_-_35201482 | 0.83 |
ENSDART00000166272
|
fcho2
|
FCH domain only 2 |
| chr3_-_26806032 | 0.82 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr6_+_52212927 | 0.81 |
ENSDART00000143458
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr19_-_31035155 | 0.80 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_-_31035325 | 0.80 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_-_340641 | 0.79 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr2_-_7800702 | 0.77 |
ENSDART00000146360
|
tbl1xr1b
|
transducin (beta)-like 1 X-linked receptor 1b |
| chr2_-_7185460 | 0.76 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr21_+_39462520 | 0.76 |
ENSDART00000114825
|
mntb
|
MAX network transcriptional repressor b |
| chr18_+_13248956 | 0.76 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr3_-_15475067 | 0.73 |
ENSDART00000025324
ENSDART00000139575 |
spns1
|
spinster homolog 1 (Drosophila) |
| chr22_-_3275888 | 0.73 |
ENSDART00000164743
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr6_+_4255319 | 0.72 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr10_-_32610776 | 0.71 |
ENSDART00000017436
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr21_+_31253048 | 0.70 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr8_+_36570791 | 0.70 |
ENSDART00000145566
ENSDART00000180527 |
pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr16_+_19637384 | 0.70 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr6_+_6779114 | 0.70 |
ENSDART00000163493
ENSDART00000143359 |
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr16_+_7991274 | 0.69 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr3_+_46764278 | 0.69 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr3_+_59051503 | 0.69 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr2_+_43204919 | 0.69 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr5_+_26204561 | 0.69 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr10_-_1961930 | 0.68 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr5_+_63329608 | 0.67 |
ENSDART00000139180
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr21_+_26522571 | 0.67 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr18_+_49969568 | 0.67 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr16_+_52999778 | 0.66 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr10_-_1961576 | 0.66 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr2_-_17393971 | 0.66 |
ENSDART00000100201
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr3_-_31716157 | 0.65 |
ENSDART00000193189
|
ccdc47
|
coiled-coil domain containing 47 |
| chr21_+_20903244 | 0.65 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr24_-_36238054 | 0.64 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr6_+_45692026 | 0.64 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr19_+_21362553 | 0.64 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr18_+_30998472 | 0.64 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr19_+_9113932 | 0.63 |
ENSDART00000060442
|
setdb1a
|
SET domain, bifurcated 1a |
| chr13_-_38039871 | 0.63 |
ENSDART00000140645
|
CR456624.1
|
|
| chr19_+_15485287 | 0.63 |
ENSDART00000182797
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr13_+_24396666 | 0.63 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
| chr14_-_26704829 | 0.62 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr3_-_40976288 | 0.62 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr13_-_30996072 | 0.62 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr9_+_2574122 | 0.62 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr23_-_29505645 | 0.62 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr10_+_11265387 | 0.60 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr3_+_33345348 | 0.60 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr8_+_36570508 | 0.60 |
ENSDART00000145868
|
pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr19_+_9232676 | 0.60 |
ENSDART00000136957
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr3_-_31715714 | 0.59 |
ENSDART00000051542
|
ccdc47
|
coiled-coil domain containing 47 |
| chr6_-_52644182 | 0.58 |
ENSDART00000015051
|
gdf5
|
growth differentiation factor 5 |
| chr6_+_19950107 | 0.58 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr3_-_25148047 | 0.57 |
ENSDART00000089325
|
mief1
|
mitochondrial elongation factor 1 |
| chr12_+_36428052 | 0.57 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr5_+_63302660 | 0.57 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr21_-_41070182 | 0.56 |
ENSDART00000026064
|
larsb
|
leucyl-tRNA synthetase b |
| chr24_+_5811808 | 0.56 |
ENSDART00000132428
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr5_-_57879138 | 0.56 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr13_+_45582391 | 0.56 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr11_+_11152214 | 0.56 |
ENSDART00000148030
|
ly75
|
lymphocyte antigen 75 |
| chr22_-_14161309 | 0.56 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr18_+_3579829 | 0.55 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr15_+_31590224 | 0.54 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr18_+_25546227 | 0.54 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr9_-_49964810 | 0.54 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr4_+_77957611 | 0.54 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr16_+_38167883 | 0.53 |
ENSDART00000111796
|
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr5_-_23749348 | 0.53 |
ENSDART00000140766
|
gbgt1l2
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 2 |
| chr10_+_2876378 | 0.53 |
ENSDART00000192083
|
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr20_-_2361226 | 0.52 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr5_+_26131690 | 0.52 |
ENSDART00000178021
ENSDART00000134371 |
si:ch211-214j8.12
|
si:ch211-214j8.12 |
| chr4_-_3353595 | 0.52 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr16_+_25107344 | 0.52 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr15_+_34592215 | 0.49 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr19_+_3140313 | 0.49 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr2_-_38206331 | 0.49 |
ENSDART00000136082
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr12_-_35582683 | 0.49 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr24_+_5811350 | 0.49 |
ENSDART00000178238
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr3_+_41558682 | 0.49 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr21_-_34926619 | 0.48 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr13_+_45476181 | 0.48 |
ENSDART00000045329
|
mgst3b
|
microsomal glutathione S-transferase 3b |
| chr25_-_31763897 | 0.48 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr16_-_43011470 | 0.48 |
ENSDART00000131898
ENSDART00000142003 ENSDART00000017966 |
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr3_+_1107102 | 0.48 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr1_+_24557414 | 0.47 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr9_+_17971935 | 0.47 |
ENSDART00000149736
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr7_-_57637779 | 0.46 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr19_+_28187480 | 0.46 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr17_-_27382826 | 0.45 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr10_+_2876020 | 0.45 |
ENSDART00000136618
ENSDART00000133128 ENSDART00000140803 ENSDART00000141505 |
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr5_+_29726428 | 0.45 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr5_-_40024902 | 0.45 |
ENSDART00000017451
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr8_+_7315625 | 0.44 |
ENSDART00000135655
ENSDART00000181048 |
selenoh
|
selenoprotein H |
| chr24_-_18809433 | 0.44 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr18_-_42830563 | 0.43 |
ENSDART00000191488
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr7_+_30725473 | 0.43 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr16_-_26125304 | 0.43 |
ENSDART00000176517
|
lipeb
|
lipase, hormone-sensitive b |
| chr24_+_8904135 | 0.43 |
ENSDART00000066782
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr13_+_24022963 | 0.42 |
ENSDART00000028285
|
pgbd5
|
piggyBac transposable element derived 5 |
| chr16_-_25380903 | 0.42 |
ENSDART00000086375
ENSDART00000188587 |
adnp2a
|
ADNP homeobox 2a |
| chr11_-_29082175 | 0.41 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr21_+_41743493 | 0.41 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr21_+_26733529 | 0.41 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr23_-_29505463 | 0.40 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr24_+_16393302 | 0.40 |
ENSDART00000188670
ENSDART00000081759 ENSDART00000177790 |
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr9_-_27738110 | 0.40 |
ENSDART00000060347
|
crygs2
|
crystallin, gamma S2 |
| chr7_+_20917966 | 0.40 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr18_+_6866276 | 0.39 |
ENSDART00000187516
|
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr11_+_18612421 | 0.39 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr5_+_26686279 | 0.39 |
ENSDART00000193543
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr4_+_5156117 | 0.39 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr2_+_35806672 | 0.38 |
ENSDART00000137384
|
rasal2
|
RAS protein activator like 2 |
| chr12_+_48480632 | 0.38 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr18_-_7481036 | 0.38 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr7_-_8315179 | 0.37 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr19_+_19976990 | 0.37 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr3_-_58165254 | 0.37 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr19_+_40145400 | 0.37 |
ENSDART00000049968
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr25_+_5604512 | 0.36 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr21_-_13662237 | 0.36 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr3_+_431208 | 0.36 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr10_+_44940693 | 0.36 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 1.0 | 5.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 1.0 | 8.2 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.5 | 2.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.5 | 1.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.5 | 1.8 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.4 | 1.3 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.4 | 1.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 1.0 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.3 | 1.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.3 | 0.9 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.3 | 1.1 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 1.0 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 0.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 2.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 0.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 0.8 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.2 | 0.8 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 0.5 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.5 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.6 | GO:0048909 | anterior lateral line nerve development(GO:0048909) |
| 0.1 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.5 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.4 | GO:0060986 | regulation of endocrine process(GO:0044060) endocrine hormone secretion(GO:0060986) |
| 0.1 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.4 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.3 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.7 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.9 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.7 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.4 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.5 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:1904867 | positive regulation of protein localization to nucleus(GO:1900182) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 1.0 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.7 | GO:0010324 | membrane invagination(GO:0010324) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.4 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.8 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.7 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.2 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.9 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.2 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.1 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.5 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.5 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 1.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 1.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.8 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 7.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.0 | 3.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 0.9 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 3.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 1.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 5.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 1.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.5 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 2.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 5.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |