Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
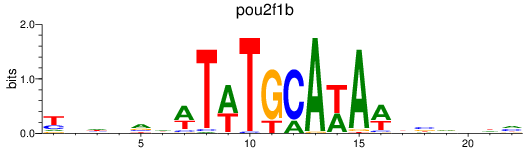
Results for pou2f1b
Z-value: 0.28
Transcription factors associated with pou2f1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f1b
|
ENSDARG00000007996 | POU class 2 homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f1b | dr11_v1_chr9_+_34350025_34350025 | 0.60 | 9.0e-03 | Click! |
Activity profile of pou2f1b motif
Sorted Z-values of pou2f1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_453619 | 1.58 |
ENSDART00000135598
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr24_-_23758003 | 1.38 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr15_+_34988148 | 0.95 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr9_-_1702648 | 0.80 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr3_-_33422738 | 0.78 |
ENSDART00000075493
|
ccdc103
|
coiled-coil domain containing 103 |
| chr25_+_13662606 | 0.74 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr4_-_8043839 | 0.69 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr25_-_9013963 | 0.66 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr22_+_17237219 | 0.60 |
ENSDART00000090069
|
axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr10_+_26667475 | 0.58 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr24_+_17269849 | 0.56 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr4_-_15603511 | 0.55 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr16_+_52847079 | 0.55 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr21_-_37733571 | 0.54 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr16_-_29557338 | 0.54 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr4_+_12013043 | 0.54 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr5_+_58372164 | 0.54 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr20_+_52492192 | 0.53 |
ENSDART00000057986
|
TSTA3 (1 of many)
|
zgc:100864 |
| chr7_+_38445938 | 0.50 |
ENSDART00000173467
|
cep89
|
centrosomal protein 89 |
| chr2_+_31665836 | 0.47 |
ENSDART00000135411
ENSDART00000143914 |
si:ch211-106h4.12
|
si:ch211-106h4.12 |
| chr3_-_59981476 | 0.47 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr1_+_52735484 | 0.45 |
ENSDART00000182076
|
CABZ01021532.1
|
|
| chr7_-_6470431 | 0.45 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr21_+_7823146 | 0.45 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr11_-_2478374 | 0.43 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr10_+_39084354 | 0.42 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr12_-_124264 | 0.42 |
ENSDART00000181551
|
FO818699.1
|
|
| chr11_-_38899816 | 0.42 |
ENSDART00000141515
|
pimr127
|
Pim proto-oncogene, serine/threonine kinase, related 127 |
| chr5_+_27432958 | 0.41 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr6_+_3934738 | 0.41 |
ENSDART00000159673
|
dync1i2b
|
dynein, cytoplasmic 1, intermediate chain 2b |
| chr15_+_19838458 | 0.41 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr1_-_56080112 | 0.41 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr23_+_19813677 | 0.41 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr8_-_44238526 | 0.40 |
ENSDART00000061115
ENSDART00000132473 ENSDART00000138019 ENSDART00000133021 |
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr6_-_37469775 | 0.40 |
ENSDART00000156546
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr7_+_20344222 | 0.40 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr5_+_66353750 | 0.39 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr20_-_43533096 | 0.39 |
ENSDART00000145199
|
pimr134
|
Pim proto-oncogene, serine/threonine kinase, related 134 |
| chr18_+_2168695 | 0.39 |
ENSDART00000165855
|
dnaaf4
|
dynein axonemal assembly factor 4 |
| chr6_-_19683406 | 0.39 |
ENSDART00000158041
|
cfap52
|
cilia and flagella associated protein 52 |
| chr20_-_13640598 | 0.38 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr11_-_11471857 | 0.36 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr6_+_26314464 | 0.36 |
ENSDART00000115392
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr21_+_45502621 | 0.35 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr8_+_24745041 | 0.34 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr22_+_38301365 | 0.34 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr19_-_40186328 | 0.33 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr2_-_43124747 | 0.33 |
ENSDART00000188883
|
lrrc6
|
leucine rich repeat containing 6 |
| chr1_+_49652967 | 0.33 |
ENSDART00000191296
|
tsga10
|
testis specific, 10 |
| chr23_-_33775145 | 0.32 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr23_+_39695827 | 0.32 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr18_-_20869175 | 0.31 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr20_+_18225329 | 0.31 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr16_-_52916248 | 0.31 |
ENSDART00000113931
|
zdhhc11
|
zinc finger, DHHC-type containing 11 |
| chr7_-_27033080 | 0.29 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr15_-_34056733 | 0.29 |
ENSDART00000170130
ENSDART00000188272 |
VWDE
|
si:dkey-30e9.7 |
| chr5_+_11840905 | 0.28 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr25_-_3830272 | 0.28 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr23_-_45339439 | 0.28 |
ENSDART00000148726
|
ccdc171
|
coiled-coil domain containing 171 |
| chr13_+_22659153 | 0.28 |
ENSDART00000143906
ENSDART00000140472 ENSDART00000078877 ENSDART00000182469 |
sncga
|
synuclein, gamma a |
| chr14_+_39255437 | 0.28 |
ENSDART00000147443
|
diaph2
|
diaphanous-related formin 2 |
| chr3_+_39759130 | 0.28 |
ENSDART00000185202
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr4_-_5831522 | 0.27 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr6_+_26309968 | 0.27 |
ENSDART00000153805
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr7_-_51102479 | 0.27 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr19_-_31584444 | 0.27 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr20_-_1383916 | 0.27 |
ENSDART00000152373
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr3_+_26135502 | 0.26 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr17_+_46864116 | 0.25 |
ENSDART00000156250
|
pimr27
|
Pim proto-oncogene, serine/threonine kinase, related 27 |
| chr2_+_5563077 | 0.25 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr14_+_33329761 | 0.25 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr10_+_33982010 | 0.25 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr18_+_19648275 | 0.25 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr18_+_26422124 | 0.25 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr7_+_19600262 | 0.24 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr3_-_59981162 | 0.24 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr6_+_2093206 | 0.24 |
ENSDART00000114314
|
tgm2b
|
transglutaminase 2b |
| chr4_-_9592402 | 0.24 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_+_8662530 | 0.24 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr6_+_35362225 | 0.24 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr1_-_51606552 | 0.23 |
ENSDART00000130828
|
cnrip1a
|
cannabinoid receptor interacting protein 1a |
| chr15_-_34558579 | 0.23 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr7_-_12968689 | 0.23 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr1_-_51615437 | 0.23 |
ENSDART00000152185
ENSDART00000152237 ENSDART00000129052 ENSDART00000152595 |
FBXO48
|
si:dkey-202b17.4 |
| chr11_-_38928760 | 0.23 |
ENSDART00000146277
|
si:ch211-122l14.6
|
si:ch211-122l14.6 |
| chr14_+_30730749 | 0.23 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr18_+_40617717 | 0.22 |
ENSDART00000049423
|
zgc:158427
|
zgc:158427 |
| chr2_-_16565690 | 0.22 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr21_+_45502773 | 0.22 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr1_+_7546259 | 0.22 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr24_+_9612218 | 0.22 |
ENSDART00000175671
|
nphp3
|
nephronophthisis 3 |
| chr5_+_58397646 | 0.22 |
ENSDART00000180759
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr19_+_1688727 | 0.22 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr25_+_31868268 | 0.21 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr20_-_45812144 | 0.21 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr6_+_23935045 | 0.21 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr19_+_43297546 | 0.21 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr8_+_26565512 | 0.21 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr6_-_442163 | 0.21 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr21_+_18877130 | 0.21 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr24_-_12770357 | 0.20 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr21_-_37733287 | 0.20 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr15_-_14098522 | 0.20 |
ENSDART00000156947
|
si:ch211-116o3.5
|
si:ch211-116o3.5 |
| chr7_-_18168493 | 0.20 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr6_-_8295657 | 0.20 |
ENSDART00000185135
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr15_-_34001000 | 0.20 |
ENSDART00000180464
|
VWDE
|
si:dkey-30e9.7 |
| chr6_+_2093367 | 0.20 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr24_+_39158283 | 0.19 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr9_-_47472998 | 0.19 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr10_+_35257651 | 0.19 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr23_+_28494189 | 0.19 |
ENSDART00000146990
ENSDART00000006657 |
huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr10_+_28587446 | 0.19 |
ENSDART00000030138
ENSDART00000137508 |
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr18_+_7639401 | 0.19 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr20_-_50049099 | 0.19 |
ENSDART00000123634
|
NDUFB1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr20_-_29052391 | 0.19 |
ENSDART00000193482
ENSDART00000017090 |
thbs1b
|
thrombospondin 1b |
| chr4_+_7827261 | 0.19 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr20_+_10544100 | 0.18 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr5_+_35458190 | 0.18 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr24_+_23791758 | 0.18 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr1_-_18803919 | 0.18 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr21_+_44013487 | 0.18 |
ENSDART00000012396
|
ik
|
IK cytokine |
| chr6_-_3573716 | 0.18 |
ENSDART00000026693
|
dmbx1b
|
diencephalon/mesencephalon homeobox 1b |
| chr7_+_17255705 | 0.18 |
ENSDART00000053357
ENSDART00000167849 ENSDART00000165833 |
nitr9
si:ch73-46n24.1
|
novel immune-type receptor 9 si:ch73-46n24.1 |
| chr1_-_10071422 | 0.17 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr2_+_45068366 | 0.17 |
ENSDART00000142175
|
si:dkey-76d14.2
|
si:dkey-76d14.2 |
| chr15_+_1134870 | 0.17 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr6_-_17112151 | 0.17 |
ENSDART00000153546
|
pimr17
|
Pim proto-oncogene, serine/threonine kinase, related 17 |
| chr4_+_12348568 | 0.17 |
ENSDART00000113595
|
pimr170
|
Pim proto-oncogene, serine/threonine kinase, related 170 |
| chr25_-_18454016 | 0.17 |
ENSDART00000005877
|
cpa1
|
carboxypeptidase A1 (pancreatic) |
| chr6_-_39764995 | 0.17 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr20_+_10538025 | 0.17 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr24_+_23959487 | 0.17 |
ENSDART00000080487
|
hgd
|
homogentisate 1,2-dioxygenase |
| chr3_+_45687266 | 0.17 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr10_+_15777258 | 0.16 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr22_+_30009926 | 0.16 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr20_+_320415 | 0.16 |
ENSDART00000152344
|
si:dkey-119m7.8
|
si:dkey-119m7.8 |
| chr16_-_45288599 | 0.16 |
ENSDART00000044097
|
hpn
|
hepsin |
| chr4_-_9764767 | 0.16 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr22_+_14051894 | 0.16 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr13_-_49802194 | 0.16 |
ENSDART00000148722
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr6_-_37468971 | 0.16 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr8_+_23436411 | 0.16 |
ENSDART00000140044
|
foxp3b
|
forkhead box P3b |
| chr15_-_43873005 | 0.16 |
ENSDART00000190326
|
NOX4
|
NADPH oxidase 4 |
| chr22_-_17585618 | 0.15 |
ENSDART00000183123
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr7_-_52531252 | 0.15 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr9_-_42730672 | 0.15 |
ENSDART00000136728
|
fkbp7
|
FK506 binding protein 7 |
| chr11_+_26476153 | 0.15 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr15_-_6863317 | 0.15 |
ENSDART00000155128
ENSDART00000018945 |
meis3
|
myeloid ecotropic viral integration site 3 |
| chr23_+_32011768 | 0.15 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr5_+_30680804 | 0.15 |
ENSDART00000078049
|
pdzd3b
|
PDZ domain containing 3b |
| chr20_+_12702923 | 0.15 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr22_-_13042992 | 0.15 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr13_+_21862050 | 0.15 |
ENSDART00000133108
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr5_-_32274383 | 0.15 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr16_-_43971258 | 0.15 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr10_+_15777064 | 0.15 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr15_+_32711663 | 0.15 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr6_+_29410986 | 0.15 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr4_-_49312747 | 0.15 |
ENSDART00000181057
|
BX942819.2
|
|
| chr10_+_42733210 | 0.14 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr20_-_39735952 | 0.14 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr12_-_6551681 | 0.14 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr21_+_8533533 | 0.14 |
ENSDART00000077924
|
FO834888.1
|
|
| chr12_+_15622621 | 0.14 |
ENSDART00000079784
|
plcd3b
|
phospholipase C, delta 3b |
| chr9_-_23891102 | 0.14 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr4_+_67050047 | 0.14 |
ENSDART00000170257
|
znf1070
|
zinc finger protein 1070 |
| chr3_-_6035766 | 0.14 |
ENSDART00000081796
|
mcoln1b
|
mucolipin 1b |
| chr21_-_20725853 | 0.14 |
ENSDART00000114502
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr3_-_18792492 | 0.14 |
ENSDART00000134208
ENSDART00000034373 |
hagh
|
hydroxyacylglutathione hydrolase |
| chr15_-_23814330 | 0.14 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr13_+_32636197 | 0.14 |
ENSDART00000141057
|
si:ch211-206k20.5
|
si:ch211-206k20.5 |
| chr3_+_46635527 | 0.14 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr9_+_32931625 | 0.13 |
ENSDART00000143484
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr19_+_4990320 | 0.13 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr4_+_9669717 | 0.13 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr11_-_29658396 | 0.13 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr11_-_38914265 | 0.13 |
ENSDART00000141229
|
si:ch211-122l14.4
|
si:ch211-122l14.4 |
| chr13_-_12006007 | 0.13 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr4_-_8035520 | 0.13 |
ENSDART00000146146
|
si:ch211-240l19.7
|
si:ch211-240l19.7 |
| chr9_-_34396264 | 0.13 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr16_-_14534724 | 0.13 |
ENSDART00000148126
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
| chr23_+_43489182 | 0.13 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr20_+_46492175 | 0.13 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr8_+_41037541 | 0.13 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr18_+_38755023 | 0.12 |
ENSDART00000010177
ENSDART00000193136 |
onecut1
|
one cut homeobox 1 |
| chr25_+_10793478 | 0.12 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr1_+_15216988 | 0.12 |
ENSDART00000189206
ENSDART00000181639 |
itln1
|
intelectin 1 |
| chr9_+_32073606 | 0.12 |
ENSDART00000184170
ENSDART00000180355 ENSDART00000110204 ENSDART00000123278 |
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr3_+_37112693 | 0.12 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr4_+_2269815 | 0.12 |
ENSDART00000075788
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr11_-_34065718 | 0.12 |
ENSDART00000110608
|
col6a1
|
collagen, type VI, alpha 1 |
| chr15_+_36187434 | 0.12 |
ENSDART00000181536
ENSDART00000099501 ENSDART00000154432 |
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr21_+_36623162 | 0.12 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr11_-_2131280 | 0.12 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr7_-_15830251 | 0.12 |
ENSDART00000102363
|
rcn1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr6_-_31576397 | 0.12 |
ENSDART00000111837
|
raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr15_-_30815826 | 0.11 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr6_+_11829867 | 0.11 |
ENSDART00000151044
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr18_-_3464222 | 0.11 |
ENSDART00000166980
|
myo7aa
|
myosin VIIAa |
| chr7_+_19495379 | 0.11 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 1.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 0.5 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.1 | 0.4 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 0.7 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.4 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.3 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.4 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.4 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.4 | GO:0061333 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.1 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.3 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.2 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.2 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.0 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0046323 | glucose import(GO:0046323) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.1 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.3 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.2 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.1 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.0 | 0.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |