Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
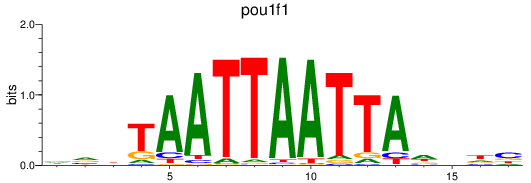
Results for pou1f1
Z-value: 0.65
Transcription factors associated with pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou1f1
|
ENSDARG00000058924 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000110816 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000115109 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou1f1 | dr11_v1_chr9_-_19161982_19161982 | 0.28 | 2.6e-01 | Click! |
Activity profile of pou1f1 motif
Sorted Z-values of pou1f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_20939579 | 1.66 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr2_+_49799470 | 1.61 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr8_-_13046089 | 1.51 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr3_+_26145013 | 1.36 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr23_+_19813677 | 1.33 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr8_-_44586981 | 1.31 |
ENSDART00000026831
ENSDART00000113945 |
rsph14
|
radial spoke head 14 homolog |
| chr5_-_54672763 | 1.27 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr12_-_35944654 | 1.23 |
ENSDART00000162579
ENSDART00000164199 |
dnai2a
|
dynein, axonemal, intermediate chain 2a |
| chr15_-_18162647 | 1.21 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr4_+_9669717 | 1.18 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr5_+_42393896 | 1.13 |
ENSDART00000189550
|
BX548073.13
|
|
| chr24_-_26328721 | 1.12 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr23_+_45512825 | 1.12 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr3_-_19200571 | 1.12 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr7_+_23515966 | 1.09 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr6_+_9677420 | 1.09 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr9_-_41062412 | 1.08 |
ENSDART00000193879
|
ankar
|
ankyrin and armadillo repeat containing |
| chr9_-_25055722 | 1.05 |
ENSDART00000137131
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr8_+_17987778 | 1.04 |
ENSDART00000133429
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr17_-_39772999 | 0.97 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr23_-_19140781 | 0.96 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr11_-_27867024 | 0.96 |
ENSDART00000182136
ENSDART00000187587 |
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr8_+_24745041 | 0.95 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr14_+_46313396 | 0.94 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_+_73397283 | 0.94 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr16_-_28856112 | 0.93 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr5_-_71705191 | 0.91 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr11_-_2838699 | 0.90 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr2_-_1548330 | 0.90 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr3_-_39180048 | 0.84 |
ENSDART00000049720
|
cdk21
|
cyclin-dependent kinase 21 |
| chr19_+_31873308 | 0.82 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr15_-_45110011 | 0.79 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr22_+_508290 | 0.78 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr6_+_40714811 | 0.78 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr3_-_12026741 | 0.78 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr5_+_69747417 | 0.77 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr8_+_17987215 | 0.75 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr13_-_37620091 | 0.75 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr23_-_23401305 | 0.75 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr24_-_38657683 | 0.74 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr1_-_46244523 | 0.71 |
ENSDART00000143908
|
si:ch211-138g9.3
|
si:ch211-138g9.3 |
| chr15_-_39955785 | 0.71 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr14_-_24761132 | 0.70 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr5_-_41494831 | 0.70 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr21_-_43015383 | 0.70 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr20_+_42881104 | 0.70 |
ENSDART00000131338
|
pimr110
|
Pim proto-oncogene, serine/threonine kinase, related 110 |
| chr20_+_19512727 | 0.69 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr7_-_71531846 | 0.69 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr3_+_36424055 | 0.69 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr21_+_45841731 | 0.67 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr10_+_26747755 | 0.66 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr23_+_12134839 | 0.66 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr14_-_1355544 | 0.66 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr2_+_14992879 | 0.65 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr9_-_7652792 | 0.65 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr20_+_25586099 | 0.64 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr16_+_28728347 | 0.64 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr2_+_19578079 | 0.64 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr13_+_4409294 | 0.63 |
ENSDART00000146437
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr12_+_13282797 | 0.61 |
ENSDART00000137757
ENSDART00000152397 |
irf9
|
interferon regulatory factor 9 |
| chr24_+_32176155 | 0.61 |
ENSDART00000003745
|
vim
|
vimentin |
| chr2_+_36004381 | 0.61 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr6_+_13117598 | 0.61 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr6_+_49551614 | 0.61 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr18_+_27821856 | 0.60 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr10_-_14556978 | 0.58 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr4_-_2545310 | 0.57 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr19_+_1688727 | 0.57 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr17_-_46817295 | 0.56 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr21_+_30502002 | 0.56 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr2_+_19522082 | 0.55 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr19_+_43297546 | 0.54 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr5_+_2815021 | 0.54 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr7_+_26534131 | 0.53 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr16_-_21140097 | 0.53 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr17_-_46933567 | 0.53 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr15_-_39945036 | 0.53 |
ENSDART00000192481
|
msh5
|
mutS homolog 5 |
| chr8_-_53044300 | 0.52 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr6_-_31361546 | 0.51 |
ENSDART00000027550
|
ak4
|
adenylate kinase 4 |
| chr15_-_46821568 | 0.51 |
ENSDART00000148525
|
chrd
|
chordin |
| chr6_+_52350443 | 0.51 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr24_-_1151334 | 0.51 |
ENSDART00000039700
ENSDART00000177356 |
itgb1a
|
integrin, beta 1a |
| chr15_+_9327252 | 0.50 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr21_-_13972745 | 0.50 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr1_-_42289704 | 0.50 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr17_+_12285285 | 0.50 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr16_+_53489676 | 0.49 |
ENSDART00000074653
|
grinab
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1b (glutamate binding) |
| chr6_+_9427641 | 0.49 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr19_-_31584444 | 0.49 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr24_+_744713 | 0.48 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr10_+_18952271 | 0.47 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr23_-_6522099 | 0.47 |
ENSDART00000092214
ENSDART00000183380 ENSDART00000138020 |
bmp7b
|
bone morphogenetic protein 7b |
| chr25_-_4146947 | 0.46 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr20_+_40457599 | 0.45 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr2_+_19578446 | 0.45 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr5_+_41322783 | 0.44 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr23_-_16485190 | 0.44 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr2_+_44571200 | 0.42 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr1_-_9980765 | 0.42 |
ENSDART00000142906
|
si:dkeyp-75b4.7
|
si:dkeyp-75b4.7 |
| chr21_+_18877130 | 0.42 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr13_-_31452516 | 0.41 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr21_-_14811058 | 0.41 |
ENSDART00000143100
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr6_-_17112151 | 0.41 |
ENSDART00000153546
|
pimr17
|
Pim proto-oncogene, serine/threonine kinase, related 17 |
| chr9_+_18716485 | 0.40 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr2_+_16160906 | 0.40 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr6_-_16868834 | 0.40 |
ENSDART00000156503
|
pimr41
|
Pim proto-oncogene, serine/threonine kinase, related 41 |
| chr6_+_25257728 | 0.40 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr2_+_36007449 | 0.40 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr1_+_25801648 | 0.39 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr2_+_3986083 | 0.39 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr7_+_26029672 | 0.38 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr20_-_9095105 | 0.38 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr6_-_17266221 | 0.37 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr20_+_11731039 | 0.37 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr14_-_470505 | 0.37 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr16_-_12173554 | 0.36 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr22_-_13851297 | 0.36 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr16_+_5408748 | 0.36 |
ENSDART00000160008
ENSDART00000014024 |
plecb
|
plectin b |
| chr4_-_948776 | 0.36 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr23_+_7710447 | 0.36 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr3_+_30922947 | 0.36 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr23_+_31405497 | 0.36 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr16_-_27566552 | 0.36 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr2_-_42065069 | 0.36 |
ENSDART00000140188
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr22_-_31060579 | 0.36 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr7_+_19495905 | 0.35 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr16_-_30434279 | 0.35 |
ENSDART00000018504
|
zgc:77086
|
zgc:77086 |
| chr16_+_18534834 | 0.35 |
ENSDART00000180168
|
rxrbb
|
retinoid x receptor, beta b |
| chr11_+_30057762 | 0.35 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr6_-_16735402 | 0.34 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr5_+_21144269 | 0.34 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr12_-_28363111 | 0.34 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr19_-_20403507 | 0.34 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr22_+_34663328 | 0.33 |
ENSDART00000183912
|
si:ch73-304f21.1
|
si:ch73-304f21.1 |
| chr4_-_2219705 | 0.33 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr12_+_22560067 | 0.33 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr12_+_3022882 | 0.33 |
ENSDART00000122905
|
rac3b
|
Rac family small GTPase 3b |
| chr19_+_43359075 | 0.33 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr19_-_20403845 | 0.32 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr9_+_28598577 | 0.32 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr20_+_41756996 | 0.32 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr7_+_59020972 | 0.32 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr25_-_4525081 | 0.32 |
ENSDART00000184347
|
pidd1
|
p53-induced death domain protein 1 |
| chr15_-_21014270 | 0.31 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr3_+_30500968 | 0.31 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr6_-_17052116 | 0.31 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr3_-_48716422 | 0.31 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr14_-_24391424 | 0.30 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr10_-_26766780 | 0.30 |
ENSDART00000146666
|
mcf2b
|
MCF.2 cell line derived transforming sequence b |
| chr11_+_40812590 | 0.30 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr8_-_17987547 | 0.29 |
ENSDART00000112699
ENSDART00000061747 |
fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr16_-_12173399 | 0.29 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr23_-_18913032 | 0.29 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr14_-_413273 | 0.29 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr1_+_15204633 | 0.29 |
ENSDART00000188410
|
itln1
|
intelectin 1 |
| chr20_+_4060839 | 0.28 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr24_-_21847945 | 0.28 |
ENSDART00000189097
|
CR352265.4
|
|
| chr16_+_46111849 | 0.28 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr6_+_11989537 | 0.27 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr10_+_2742499 | 0.27 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr25_-_37186894 | 0.27 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr9_-_40765868 | 0.27 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr8_+_28259347 | 0.26 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr11_+_45436703 | 0.26 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr3_+_38540411 | 0.26 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr10_+_44824072 | 0.26 |
ENSDART00000162669
|
slc4a5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr12_+_47698356 | 0.26 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr21_+_45502773 | 0.26 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr1_+_49668423 | 0.26 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr21_-_13123176 | 0.26 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr7_+_71664624 | 0.25 |
ENSDART00000170273
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr8_+_52637507 | 0.25 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr5_-_2282256 | 0.25 |
ENSDART00000064012
|
ca4a
|
carbonic anhydrase IV a |
| chr7_+_19495379 | 0.25 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr3_-_15999501 | 0.25 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr21_+_25236297 | 0.25 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr6_-_50704689 | 0.25 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr14_-_31854830 | 0.24 |
ENSDART00000148550
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr2_-_9059955 | 0.24 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr17_-_29224908 | 0.24 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr14_+_38845674 | 0.24 |
ENSDART00000129958
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr16_+_54209504 | 0.24 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr19_-_26869103 | 0.24 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr13_+_29770837 | 0.23 |
ENSDART00000076998
|
pax2a
|
paired box 2a |
| chr25_+_28825657 | 0.23 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr2_+_16487443 | 0.23 |
ENSDART00000114980
|
CR377211.1
|
|
| chr22_-_5252005 | 0.23 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr25_+_14087045 | 0.23 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr16_+_28994709 | 0.23 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr9_+_29548195 | 0.23 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr6_-_17179637 | 0.23 |
ENSDART00000153710
|
pimr19
|
Pim proto-oncogene, serine/threonine kinase, related 19 |
| chr11_-_29737088 | 0.22 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr23_+_45200481 | 0.22 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr8_+_17775247 | 0.22 |
ENSDART00000112356
|
si:ch211-150o23.3
|
si:ch211-150o23.3 |
| chr15_-_40267485 | 0.22 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr8_-_25569920 | 0.21 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr2_+_42072231 | 0.21 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr6_-_43283122 | 0.21 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr23_-_18024543 | 0.21 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr24_+_9475809 | 0.21 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou1f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 1.4 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.2 | 1.4 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 0.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 1.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.4 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.8 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.6 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 1.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.9 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.5 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.4 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.6 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.2 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.2 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.4 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 1.1 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 1.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0060118 | cerebellum formation(GO:0021588) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 9.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.5 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.9 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.9 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.5 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0045730 | respiratory burst(GO:0045730) defense response to fungus(GO:0050832) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.5 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 1.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.4 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.5 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.6 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 1.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 1.7 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |