Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for pitx2
Z-value: 0.59
Transcription factors associated with pitx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pitx2
|
ENSDARG00000036194 | paired-like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pitx2 | dr11_v1_chr14_+_36218072_36218072 | -0.22 | 3.8e-01 | Click! |
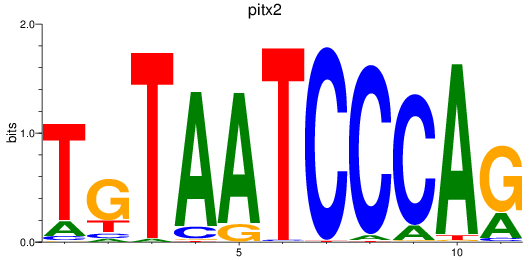
Activity profile of pitx2 motif
Sorted Z-values of pitx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_33216945 | 1.10 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr2_+_6253246 | 1.08 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr24_-_10006158 | 0.97 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr7_+_24114694 | 0.88 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr7_+_24115082 | 0.83 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr19_+_28284120 | 0.74 |
ENSDART00000103862
|
mrpl36
|
mitochondrial ribosomal protein L36 |
| chr2_-_3419890 | 0.67 |
ENSDART00000055618
|
iba57
|
IBA57, iron-sulfur cluster assembly |
| chr3_-_3448095 | 0.63 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr2_+_37295088 | 0.62 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr15_-_8319988 | 0.60 |
ENSDART00000137552
|
si:ch211-113p18.3
|
si:ch211-113p18.3 |
| chr1_-_8651718 | 0.57 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr15_+_37545855 | 0.54 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr10_-_43771447 | 0.51 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr1_+_55755304 | 0.51 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr3_-_40528333 | 0.50 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr8_-_21372446 | 0.50 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr12_+_48390715 | 0.49 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr9_+_14028157 | 0.49 |
ENSDART00000134603
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr9_-_23747264 | 0.47 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr5_-_15948833 | 0.45 |
ENSDART00000051649
ENSDART00000124467 |
xbp1
|
X-box binding protein 1 |
| chr19_-_40186328 | 0.43 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr4_+_11464255 | 0.43 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr9_+_1365747 | 0.42 |
ENSDART00000140917
ENSDART00000036605 |
prkra
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr21_+_11367253 | 0.42 |
ENSDART00000091835
|
gtf3c5
|
general transcription factor IIIC, polypeptide 5 |
| chr12_+_17106117 | 0.39 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr4_-_73256421 | 0.36 |
ENSDART00000174158
|
CABZ01021433.1
|
|
| chr5_+_13891202 | 0.36 |
ENSDART00000193992
|
hk2
|
hexokinase 2 |
| chr5_+_62052538 | 0.35 |
ENSDART00000141574
|
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr18_+_20482369 | 0.35 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr5_+_62052750 | 0.35 |
ENSDART00000192103
ENSDART00000181866 |
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr9_+_34127005 | 0.34 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr10_+_14982977 | 0.33 |
ENSDART00000140869
|
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr8_+_27807974 | 0.33 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr25_-_17357165 | 0.32 |
ENSDART00000064573
|
setd6
|
SET domain containing 6 |
| chr14_-_15699528 | 0.32 |
ENSDART00000161123
|
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr19_+_19989380 | 0.30 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr18_-_12270845 | 0.30 |
ENSDART00000143923
ENSDART00000125555 ENSDART00000062360 ENSDART00000062216 |
nup205
|
nucleoporin 205 |
| chr16_+_33902006 | 0.30 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr5_+_37749503 | 0.30 |
ENSDART00000148586
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr15_-_25584888 | 0.29 |
ENSDART00000127571
|
si:dkey-54n8.2
|
si:dkey-54n8.2 |
| chr1_-_57172294 | 0.29 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr16_-_30847192 | 0.29 |
ENSDART00000191106
ENSDART00000128158 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr10_-_45029041 | 0.28 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr5_-_20491198 | 0.28 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr15_+_37546391 | 0.26 |
ENSDART00000186625
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr13_+_43400443 | 0.26 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr5_-_23117078 | 0.26 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr5_+_58687541 | 0.26 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr11_+_5926850 | 0.25 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr22_-_18491813 | 0.25 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr16_-_17258111 | 0.25 |
ENSDART00000079497
|
emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr11_-_36051004 | 0.25 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr17_-_53328043 | 0.24 |
ENSDART00000171082
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr14_+_44545092 | 0.24 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr1_+_7546259 | 0.24 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr25_-_26833100 | 0.24 |
ENSDART00000014052
|
neil1
|
nei-like DNA glycosylase 1 |
| chr9_+_22631672 | 0.24 |
ENSDART00000101770
ENSDART00000126015 ENSDART00000145005 |
etv5a
|
ets variant 5a |
| chr6_-_21091948 | 0.23 |
ENSDART00000057348
|
inha
|
inhibin, alpha |
| chr8_-_54304381 | 0.23 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr13_+_24280380 | 0.22 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr14_+_555674 | 0.21 |
ENSDART00000172137
|
nudt6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr12_+_47917971 | 0.21 |
ENSDART00000185933
|
tbata
|
thymus, brain and testes associated |
| chr23_+_44349252 | 0.21 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr7_-_19369002 | 0.21 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr22_-_9889413 | 0.21 |
ENSDART00000081450
|
si:dkey-253d23.5
|
si:dkey-253d23.5 |
| chr20_+_263056 | 0.21 |
ENSDART00000132669
|
tube1
|
tubulin, epsilon 1 |
| chr19_-_28283844 | 0.20 |
ENSDART00000151756
ENSDART00000079104 |
ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr12_+_48216662 | 0.20 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr9_+_22632126 | 0.20 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr4_-_9579299 | 0.20 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr11_-_26375575 | 0.20 |
ENSDART00000079255
|
utp3
|
UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
| chr14_-_897874 | 0.20 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr9_+_54695981 | 0.19 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr13_+_36958086 | 0.19 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr11_+_6456146 | 0.19 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr17_+_16046314 | 0.19 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr6_+_13083146 | 0.17 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr17_+_16046132 | 0.17 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr23_-_36313431 | 0.17 |
ENSDART00000125860
|
nfe2
|
nuclear factor, erythroid 2 |
| chr25_+_14398650 | 0.17 |
ENSDART00000130548
|
ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr19_+_34742706 | 0.17 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr15_-_20024205 | 0.17 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr17_+_24445818 | 0.17 |
ENSDART00000139963
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr7_+_38588866 | 0.16 |
ENSDART00000015682
|
ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) |
| chr25_+_25124684 | 0.16 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr1_+_33383644 | 0.16 |
ENSDART00000187194
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr20_+_50052627 | 0.16 |
ENSDART00000188799
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr6_+_2093206 | 0.15 |
ENSDART00000114314
|
tgm2b
|
transglutaminase 2b |
| chr20_+_50052116 | 0.15 |
ENSDART00000047212
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr23_+_39413163 | 0.15 |
ENSDART00000184254
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr17_-_16342388 | 0.14 |
ENSDART00000017930
|
kcnk13a
|
potassium channel, subfamily K, member 13a |
| chr21_+_13205859 | 0.14 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr1_-_44812014 | 0.14 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr25_-_35047838 | 0.14 |
ENSDART00000155060
|
CU302436.3
|
|
| chr19_-_15855427 | 0.14 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr6_-_54179860 | 0.13 |
ENSDART00000164283
|
rps10
|
ribosomal protein S10 |
| chr20_-_25369767 | 0.13 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr1_-_12394048 | 0.13 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr21_+_3854414 | 0.13 |
ENSDART00000122699
|
miga2
|
mitoguardin 2 |
| chr21_-_39177564 | 0.13 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr6_+_29217392 | 0.12 |
ENSDART00000006386
|
atp1b1a
|
ATPase Na+/K+ transporting subunit beta 1a |
| chr25_+_26844028 | 0.12 |
ENSDART00000127274
ENSDART00000156179 |
sema7a
|
semaphorin 7A |
| chr10_+_15777258 | 0.12 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr10_+_15777064 | 0.12 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr4_+_8168514 | 0.12 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr2_-_6292510 | 0.12 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr24_-_21258945 | 0.12 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr24_-_24168475 | 0.12 |
ENSDART00000139369
ENSDART00000080671 |
eif1axb
|
eukaryotic translation initiation factor 1A, X-linked, b |
| chr6_-_39903393 | 0.12 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr23_+_22267374 | 0.12 |
ENSDART00000079035
|
rap1gap
|
RAP1 GTPase activating protein |
| chr4_-_74109792 | 0.12 |
ENSDART00000174320
|
RAB21
|
zgc:154045 |
| chr20_-_20533865 | 0.11 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr23_+_10396842 | 0.11 |
ENSDART00000167593
ENSDART00000125103 |
eif4ba
|
eukaryotic translation initiation factor 4Ba |
| chr8_-_47844456 | 0.11 |
ENSDART00000145429
|
si:dkeyp-104h9.5
|
si:dkeyp-104h9.5 |
| chr11_-_18468570 | 0.11 |
ENSDART00000155474
ENSDART00000193869 |
fgd5a
|
FYVE, RhoGEF and PH domain containing 5a |
| chr16_-_21692024 | 0.11 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr5_+_26075230 | 0.11 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr3_+_37574885 | 0.11 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr1_-_43987873 | 0.11 |
ENSDART00000108821
|
CR385050.1
|
|
| chr8_-_22508055 | 0.11 |
ENSDART00000101616
|
si:ch211-261n11.5
|
si:ch211-261n11.5 |
| chr15_+_5973909 | 0.11 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr5_+_27421639 | 0.10 |
ENSDART00000146285
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr8_-_23612462 | 0.10 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr4_-_77551860 | 0.10 |
ENSDART00000188176
|
AL935186.6
|
|
| chr13_+_36984794 | 0.10 |
ENSDART00000137328
|
frmd6
|
FERM domain containing 6 |
| chr16_+_20167811 | 0.10 |
ENSDART00000004031
|
hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr1_-_37383741 | 0.10 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr9_-_53537989 | 0.10 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr14_-_32884138 | 0.09 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr11_-_270210 | 0.09 |
ENSDART00000005217
ENSDART00000172779 |
alas1
|
aminolevulinate, delta-, synthase 1 |
| chr5_+_24287927 | 0.09 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr2_-_2096055 | 0.09 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr7_+_828289 | 0.09 |
ENSDART00000173796
|
cabz01076234.2
|
cabz01076234.2 |
| chr1_-_8966046 | 0.09 |
ENSDART00000136198
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr7_-_66864756 | 0.09 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr12_+_30706158 | 0.09 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr3_+_52806347 | 0.09 |
ENSDART00000168151
ENSDART00000098826 |
lmx1al
|
LIM homeobox transcription factor 1, alpha-like |
| chr25_+_35019693 | 0.09 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr12_+_28995942 | 0.09 |
ENSDART00000076334
|
valopb
|
vertebrate ancient long opsin b |
| chr14_-_40821411 | 0.08 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr11_-_1933793 | 0.08 |
ENSDART00000186850
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr6_-_11812224 | 0.08 |
ENSDART00000150989
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr23_+_9220436 | 0.08 |
ENSDART00000033663
ENSDART00000139870 |
rps21
|
ribosomal protein S21 |
| chr16_-_50952266 | 0.08 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr19_+_12762887 | 0.08 |
ENSDART00000139909
|
mc5ra
|
melanocortin 5a receptor |
| chr11_+_6115621 | 0.08 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr8_+_694218 | 0.08 |
ENSDART00000147753
|
rnf165b
|
ring finger protein 165b |
| chr14_-_2004291 | 0.08 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr1_-_37383539 | 0.08 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr5_-_26493253 | 0.08 |
ENSDART00000088222
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr14_+_6423973 | 0.08 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr23_-_15330168 | 0.08 |
ENSDART00000035865
ENSDART00000143635 |
sulf2b
|
sulfatase 2b |
| chr3_-_7940522 | 0.08 |
ENSDART00000159710
ENSDART00000167201 |
trim35-25
|
tripartite motif containing 35-25 |
| chr11_-_28148033 | 0.08 |
ENSDART00000177182
|
lactbl1b
|
lactamase, beta-like 1b |
| chr21_-_22715297 | 0.08 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr8_+_17184602 | 0.07 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr14_-_2270973 | 0.07 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr7_+_37377335 | 0.07 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr17_-_31659670 | 0.07 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr18_+_507835 | 0.07 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr11_+_6116096 | 0.07 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr12_+_316238 | 0.07 |
ENSDART00000187492
|
rcvrnb
|
recoverin b |
| chr10_+_26926654 | 0.07 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr10_-_7857494 | 0.07 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr11_+_6116503 | 0.07 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr7_+_47316944 | 0.07 |
ENSDART00000173534
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr17_-_50331351 | 0.07 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr4_-_1403077 | 0.07 |
ENSDART00000189813
|
CABZ01068469.1
|
|
| chr1_-_10577945 | 0.06 |
ENSDART00000179237
ENSDART00000040502 ENSDART00000186876 |
trpc5a
|
transient receptor potential cation channel, subfamily C, member 5a |
| chr21_+_19648814 | 0.06 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr20_-_30369598 | 0.06 |
ENSDART00000144549
|
allc
|
allantoicase |
| chr4_-_40883162 | 0.06 |
ENSDART00000151978
|
znf1102
|
zinc finger protein 1102 |
| chr2_-_2020044 | 0.06 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr4_+_28442223 | 0.06 |
ENSDART00000150879
|
znf1067
|
zinc finger protein 1067 |
| chr21_+_40944530 | 0.06 |
ENSDART00000022976
|
kctd16b
|
potassium channel tetramerization domain containing 16b |
| chr10_+_29137482 | 0.06 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr23_+_8989168 | 0.06 |
ENSDART00000034380
|
xkr7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr16_+_3067134 | 0.06 |
ENSDART00000012048
|
cyb5r4
|
cytochrome b5 reductase 4 |
| chr22_+_696931 | 0.06 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr6_-_55354004 | 0.06 |
ENSDART00000165911
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr16_-_28836523 | 0.06 |
ENSDART00000108596
ENSDART00000142920 |
plin6
|
perilipin 6 |
| chr20_-_42534504 | 0.06 |
ENSDART00000132310
|
rfx6
|
regulatory factor X, 6 |
| chr10_-_43392267 | 0.06 |
ENSDART00000142872
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr17_-_24714837 | 0.06 |
ENSDART00000154871
|
si:ch211-15d5.11
|
si:ch211-15d5.11 |
| chr17_+_21760032 | 0.06 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr22_+_19486711 | 0.06 |
ENSDART00000144389
ENSDART00000139256 ENSDART00000160855 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr7_+_19738665 | 0.06 |
ENSDART00000089333
|
tkfc
|
triokinase/FMN cyclase |
| chr13_+_669177 | 0.06 |
ENSDART00000190085
|
CU462915.1
|
|
| chr25_+_3788074 | 0.06 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr10_-_25204034 | 0.06 |
ENSDART00000161100
|
FAT3 (1 of many)
|
si:ch211-214k5.6 |
| chr15_-_47848544 | 0.06 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr5_-_67145505 | 0.06 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr17_+_28624321 | 0.06 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr4_-_44500201 | 0.06 |
ENSDART00000150460
|
si:dkeyp-100h4.7
|
si:dkeyp-100h4.7 |
| chr20_-_54564018 | 0.06 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr17_+_44756247 | 0.06 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr23_-_24856025 | 0.06 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr4_+_5848229 | 0.06 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr14_-_3132188 | 0.06 |
ENSDART00000172259
|
si:zfos-1069f5.1
|
si:zfos-1069f5.1 |
| chr23_+_23232136 | 0.06 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pitx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 1.1 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 1.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.8 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.2 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.0 | 0.1 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.3 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 2.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 1.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.0 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.0 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |