Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for phox2bb
Z-value: 0.22
Transcription factors associated with phox2bb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2bb
|
ENSDARG00000091029 | paired like homeobox 2Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2bb | dr11_v1_chr14_-_17068712_17068712 | 0.05 | 8.3e-01 | Click! |
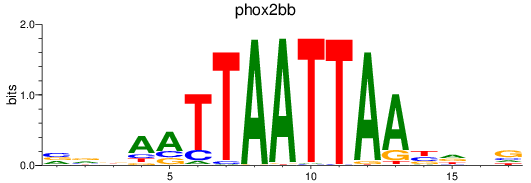
Activity profile of phox2bb motif
Sorted Z-values of phox2bb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_1801519 | 0.22 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr8_-_53044300 | 0.18 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr15_-_43284021 | 0.18 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr10_-_13343831 | 0.15 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr5_+_63302660 | 0.14 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr22_-_14247276 | 0.13 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr18_+_2228737 | 0.13 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr9_-_32177117 | 0.12 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr22_-_10156581 | 0.12 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr14_-_470505 | 0.10 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr9_-_27398369 | 0.10 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr22_+_2751887 | 0.10 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr21_+_25236297 | 0.09 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr4_-_2219705 | 0.09 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr24_+_28953089 | 0.09 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr22_+_23359369 | 0.09 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr6_+_25257728 | 0.09 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr22_+_508290 | 0.09 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr1_-_31534089 | 0.08 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr2_-_17393216 | 0.08 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr10_+_11261576 | 0.07 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr8_-_49728590 | 0.07 |
ENSDART00000135714
ENSDART00000138810 ENSDART00000098319 |
gkap1
|
G kinase anchoring protein 1 |
| chr24_+_19415124 | 0.07 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr3_-_48716422 | 0.07 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr21_-_13123176 | 0.07 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr5_+_29851433 | 0.07 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr2_-_17392799 | 0.06 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr1_+_10318089 | 0.05 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr17_-_4245902 | 0.05 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr3_-_26787430 | 0.03 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr13_+_33268657 | 0.03 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr7_+_38811800 | 0.02 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr23_+_16638639 | 0.02 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr6_+_40922572 | 0.02 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr9_+_54039006 | 0.02 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr15_+_5360407 | 0.02 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr6_+_11249706 | 0.02 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr1_-_21287724 | 0.02 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr5_+_60590796 | 0.02 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr1_-_15797663 | 0.01 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr8_-_33381913 | 0.01 |
ENSDART00000076420
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr14_-_33481428 | 0.01 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr14_-_4556896 | 0.01 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr23_-_2901167 | 0.01 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr15_-_39955785 | 0.01 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr4_-_9891874 | 0.01 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr10_-_27049170 | 0.01 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr6_+_24420523 | 0.00 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr23_-_16485190 | 0.00 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr6_+_11250033 | 0.00 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr13_+_22476742 | 0.00 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2bb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |