Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
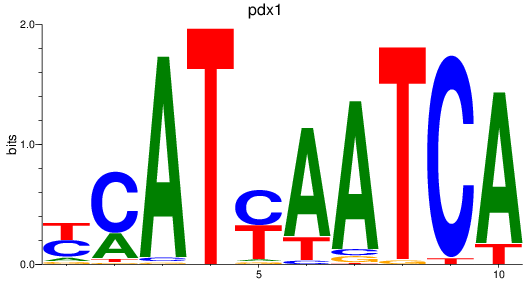
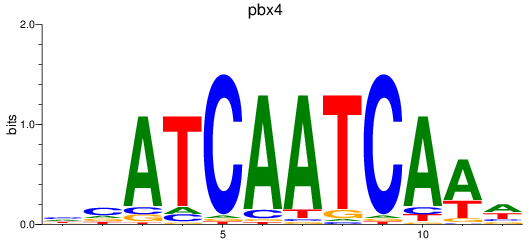
Results for pdx1_pbx4
Z-value: 0.34
Transcription factors associated with pdx1_pbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pdx1
|
ENSDARG00000002779 | pancreatic and duodenal homeobox 1 |
|
pdx1
|
ENSDARG00000109275 | pancreatic and duodenal homeobox 1 |
|
pbx4
|
ENSDARG00000052150 | pre-B-cell leukemia transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pbx4 | dr11_v1_chr3_+_53052769_53052769 | -0.35 | 1.5e-01 | Click! |
| pdx1 | dr11_v1_chr24_+_21684189_21684189 | -0.06 | 8.3e-01 | Click! |
Activity profile of pdx1_pbx4 motif
Sorted Z-values of pdx1_pbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_4461104 | 0.36 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr3_+_23752150 | 0.33 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr11_+_36243774 | 0.31 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr1_+_16144615 | 0.29 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr7_-_35066457 | 0.27 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr24_+_5789790 | 0.24 |
ENSDART00000189600
|
BX470065.1
|
|
| chr18_+_13792490 | 0.20 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr2_-_10821053 | 0.18 |
ENSDART00000056034
|
rpap2
|
RNA polymerase II associated protein 2 |
| chr17_-_2595736 | 0.18 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_6898302 | 0.17 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr6_-_23619428 | 0.17 |
ENSDART00000053126
|
aanat1
|
arylalkylamine N-acetyltransferase 1 |
| chr9_+_42066030 | 0.16 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr16_+_32736588 | 0.16 |
ENSDART00000075191
ENSDART00000168358 |
zgc:172323
|
zgc:172323 |
| chr18_-_26715655 | 0.16 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr16_+_25126935 | 0.16 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr1_-_9277986 | 0.16 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr13_+_17468161 | 0.15 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr2_-_54039293 | 0.14 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr24_+_5789582 | 0.14 |
ENSDART00000141504
|
BX470065.1
|
|
| chr10_+_45345574 | 0.14 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr11_-_25538341 | 0.14 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr9_-_3653259 | 0.14 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr24_+_13735616 | 0.14 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr3_+_18936009 | 0.14 |
ENSDART00000140330
|
si:ch211-198m1.1
|
si:ch211-198m1.1 |
| chr1_-_40227166 | 0.14 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr9_+_50000504 | 0.13 |
ENSDART00000164409
ENSDART00000165451 ENSDART00000166509 |
slc38a11
|
solute carrier family 38, member 11 |
| chr5_-_30535327 | 0.13 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr20_+_18703108 | 0.13 |
ENSDART00000181188
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr20_+_18702624 | 0.12 |
ENSDART00000185605
ENSDART00000019476 ENSDART00000152533 |
eif5
|
eukaryotic translation initiation factor 5 |
| chr6_+_3640381 | 0.12 |
ENSDART00000172078
|
col28a2b
|
collagen, type XXVIII, alpha 2b |
| chr18_-_26715156 | 0.12 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr22_+_35089031 | 0.12 |
ENSDART00000076040
|
srfa
|
serum response factor a |
| chr14_+_5383060 | 0.12 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr10_-_8046764 | 0.12 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr21_+_5192016 | 0.11 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr10_-_8053753 | 0.11 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr20_-_8443425 | 0.11 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr2_+_27855346 | 0.11 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr2_+_27855102 | 0.11 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr3_-_61116258 | 0.10 |
ENSDART00000009194
ENSDART00000156978 |
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr3_+_56366395 | 0.10 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr2_+_47623202 | 0.10 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr1_-_43963441 | 0.10 |
ENSDART00000137646
|
odam
|
odontogenic, ameloblast associated |
| chr3_-_32818607 | 0.10 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr15_+_14861335 | 0.10 |
ENSDART00000166250
|
CR936977.2
|
|
| chr18_-_42172101 | 0.09 |
ENSDART00000124211
|
cntn5
|
contactin 5 |
| chr24_-_6897884 | 0.09 |
ENSDART00000080766
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr22_-_16154771 | 0.09 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr8_+_21376290 | 0.09 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr3_-_55099272 | 0.09 |
ENSDART00000130869
|
hbba1
|
hemoglobin, beta adult 1 |
| chr2_-_21625493 | 0.09 |
ENSDART00000137169
|
zgc:55781
|
zgc:55781 |
| chr10_+_31244619 | 0.09 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr5_+_4298636 | 0.09 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr4_-_133492 | 0.09 |
ENSDART00000171845
|
gpr19
|
G protein-coupled receptor 19 |
| chr18_-_7677208 | 0.09 |
ENSDART00000092456
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr13_+_38302665 | 0.08 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr14_+_5385855 | 0.08 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr6_+_12316438 | 0.08 |
ENSDART00000156854
|
si:dkey-276j7.2
|
si:dkey-276j7.2 |
| chr23_+_28322986 | 0.08 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr1_-_56948694 | 0.08 |
ENSDART00000152597
|
si:ch211-1f22.1
|
si:ch211-1f22.1 |
| chr12_-_48970299 | 0.08 |
ENSDART00000163734
|
rgrb
|
retinal G protein coupled receptor b |
| chr12_+_17201522 | 0.08 |
ENSDART00000014536
ENSDART00000152452 |
rnls
|
renalase, FAD-dependent amine oxidase |
| chr13_+_30421472 | 0.08 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr19_+_28256076 | 0.08 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr6_-_31224563 | 0.08 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr6_-_25371196 | 0.08 |
ENSDART00000187291
|
PKN2 (1 of many)
|
zgc:153916 |
| chr10_+_76864 | 0.08 |
ENSDART00000036375
|
vps26c
|
Down syndrome critical region 3 |
| chr6_-_49510553 | 0.08 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr5_+_72377851 | 0.08 |
ENSDART00000160479
|
lman2la
|
lectin, mannose-binding 2-like a |
| chr22_-_38508112 | 0.07 |
ENSDART00000192004
|
klc4
|
kinesin light chain 4 |
| chr5_-_11573490 | 0.07 |
ENSDART00000109577
|
FO704871.1
|
|
| chr6_+_149405 | 0.07 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr8_+_10001805 | 0.07 |
ENSDART00000132894
|
si:dkey-8e10.2
|
si:dkey-8e10.2 |
| chr11_+_13528437 | 0.07 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr12_+_39685485 | 0.07 |
ENSDART00000163403
|
LO017650.1
|
|
| chr25_+_5972690 | 0.07 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr2_-_20574193 | 0.07 |
ENSDART00000190448
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr24_+_7884880 | 0.07 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr17_-_16342388 | 0.07 |
ENSDART00000017930
|
kcnk13a
|
potassium channel, subfamily K, member 13a |
| chr14_-_6666854 | 0.07 |
ENSDART00000133031
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr18_+_3572314 | 0.07 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr9_-_43644261 | 0.07 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr16_-_54498109 | 0.07 |
ENSDART00000083713
|
clk2b
|
CDC-like kinase 2b |
| chr13_-_44834895 | 0.06 |
ENSDART00000135573
|
si:dkeyp-2e4.5
|
si:dkeyp-2e4.5 |
| chr3_+_4502066 | 0.06 |
ENSDART00000088610
|
rangap1a
|
RAN GTPase activating protein 1a |
| chr14_-_23684814 | 0.06 |
ENSDART00000024604
|
mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr24_-_25144441 | 0.06 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr7_-_59564011 | 0.06 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr20_+_4793516 | 0.06 |
ENSDART00000053875
|
lgals8a
|
galectin 8a |
| chr5_-_25733745 | 0.06 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr13_+_14006118 | 0.06 |
ENSDART00000131875
ENSDART00000089528 |
atrn
|
attractin |
| chr19_+_33476557 | 0.06 |
ENSDART00000181800
|
triqk
|
triple QxxK/R motif containing |
| chr11_-_5865744 | 0.06 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr1_-_43892349 | 0.06 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr1_-_26023678 | 0.06 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr1_+_39387876 | 0.06 |
ENSDART00000084891
ENSDART00000148701 |
tenm3
|
teneurin transmembrane protein 3 |
| chr23_+_384850 | 0.06 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr24_-_2736809 | 0.06 |
ENSDART00000171805
|
fars2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr10_-_39011514 | 0.06 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr24_+_5811808 | 0.06 |
ENSDART00000132428
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr9_-_23156908 | 0.06 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr15_-_27710513 | 0.06 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr13_-_28272299 | 0.06 |
ENSDART00000006393
|
tlx1
|
T cell leukemia homeobox 1 |
| chr4_-_42408339 | 0.06 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr20_+_1088658 | 0.06 |
ENSDART00000162991
|
BX537249.1
|
|
| chr7_+_51774502 | 0.06 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr1_+_9144958 | 0.06 |
ENSDART00000046507
|
rasd3
|
RASD family member 3 |
| chr13_+_22480496 | 0.05 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr19_-_48039400 | 0.05 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr13_-_44423 | 0.05 |
ENSDART00000093247
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr12_+_2870671 | 0.05 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr21_+_31151197 | 0.05 |
ENSDART00000137728
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr4_-_43731342 | 0.05 |
ENSDART00000146627
|
si:ch211-226o13.3
|
si:ch211-226o13.3 |
| chr4_-_77561679 | 0.05 |
ENSDART00000180809
|
AL935186.9
|
|
| chr12_+_23664335 | 0.05 |
ENSDART00000111334
|
mtpap
|
mitochondrial poly(A) polymerase |
| chr5_+_45007962 | 0.05 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr23_+_4689626 | 0.05 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr2_+_2503396 | 0.05 |
ENSDART00000168418
|
crhr2
|
corticotropin releasing hormone receptor 2 |
| chr20_+_4793790 | 0.05 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr7_+_27041315 | 0.05 |
ENSDART00000052730
|
rps13
|
ribosomal protein S13 |
| chr5_+_23622177 | 0.05 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr9_+_3548529 | 0.05 |
ENSDART00000130861
ENSDART00000123074 ENSDART00000147650 |
tlk1a
|
tousled-like kinase 1a |
| chr17_+_31914877 | 0.05 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr21_-_5879897 | 0.05 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr10_-_34002185 | 0.05 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr25_+_11456696 | 0.05 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr15_-_16704417 | 0.05 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr14_+_3287740 | 0.05 |
ENSDART00000186290
|
cdx1a
|
caudal type homeobox 1a |
| chr20_-_20270191 | 0.05 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr25_+_16117138 | 0.05 |
ENSDART00000146604
|
far1
|
fatty acyl CoA reductase 1 |
| chr5_-_33287691 | 0.05 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr18_+_6126506 | 0.05 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr8_-_53488832 | 0.05 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr12_-_17201028 | 0.05 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr7_-_71384391 | 0.05 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr21_-_23307653 | 0.05 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr12_+_33975065 | 0.05 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr2_+_3809226 | 0.04 |
ENSDART00000147261
|
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr11_-_1509773 | 0.04 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr20_+_1316495 | 0.04 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr4_-_5366566 | 0.04 |
ENSDART00000150788
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr14_+_4796168 | 0.04 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr17_-_200316 | 0.04 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr12_-_26430507 | 0.04 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr15_+_31815594 | 0.04 |
ENSDART00000192525
|
frya
|
furry homolog a (Drosophila) |
| chr6_-_30689126 | 0.04 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial |
| chr8_-_38201415 | 0.04 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr8_-_32346300 | 0.04 |
ENSDART00000145325
|
ipo11
|
importin 11 |
| chr13_+_22479988 | 0.04 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr20_+_21583639 | 0.04 |
ENSDART00000131069
|
esr2a
|
estrogen receptor 2a |
| chr18_+_45889496 | 0.04 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr12_-_19091214 | 0.04 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr19_+_37925616 | 0.04 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr16_+_46869010 | 0.04 |
ENSDART00000153619
|
thsd7ab
|
thrombospondin, type I, domain containing 7Ab |
| chr14_-_30876708 | 0.04 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr7_+_12927944 | 0.04 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr7_-_51300277 | 0.04 |
ENSDART00000174174
|
gc2
|
guanylyl cyclase 2 |
| chr24_-_26820698 | 0.04 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr4_-_77331797 | 0.04 |
ENSDART00000162325
|
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr20_+_32152355 | 0.04 |
ENSDART00000152904
ENSDART00000139507 |
sesn1
|
sestrin 1 |
| chr12_-_1361517 | 0.04 |
ENSDART00000188297
|
LO018020.2
|
|
| chr14_-_45702721 | 0.04 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr4_-_77332032 | 0.04 |
ENSDART00000168628
ENSDART00000172025 |
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr4_+_44454270 | 0.04 |
ENSDART00000150482
|
si:dkeyp-100h4.4
|
si:dkeyp-100h4.4 |
| chr2_+_42724404 | 0.04 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr8_-_16697912 | 0.04 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr13_-_36621926 | 0.04 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr22_+_38914983 | 0.04 |
ENSDART00000085701
|
senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr7_-_41013575 | 0.04 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr10_+_43039947 | 0.04 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr7_-_46777876 | 0.04 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr20_-_43663494 | 0.04 |
ENSDART00000144564
|
BX470188.1
|
|
| chr2_+_17451656 | 0.04 |
ENSDART00000163620
|
BX640408.1
|
|
| chr9_-_27717006 | 0.04 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr21_+_22845317 | 0.04 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr24_-_40009446 | 0.04 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr14_-_2199573 | 0.04 |
ENSDART00000124485
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr2_-_32457919 | 0.04 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr15_-_34567370 | 0.03 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr10_-_17466990 | 0.03 |
ENSDART00000147794
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr9_+_37329036 | 0.03 |
ENSDART00000131756
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr11_-_11625369 | 0.03 |
ENSDART00000112328
|
si:dkey-28e7.3
|
si:dkey-28e7.3 |
| chr2_+_53357953 | 0.03 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr13_-_24447332 | 0.03 |
ENSDART00000043004
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr7_+_25036188 | 0.03 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr2_+_7818368 | 0.03 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr11_-_17713987 | 0.03 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr1_-_44812014 | 0.03 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr3_+_62353650 | 0.03 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr21_-_23308286 | 0.03 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr9_+_27379014 | 0.03 |
ENSDART00000167366
|
tlr20.4
|
toll-like receptor 20, tandem duplicate 4 |
| chr13_-_15700060 | 0.03 |
ENSDART00000170689
ENSDART00000010986 ENSDART00000101741 ENSDART00000139124 |
ckba
|
creatine kinase, brain a |
| chr1_+_35985813 | 0.03 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr7_-_69983948 | 0.03 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr16_+_14707960 | 0.03 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr16_+_23282655 | 0.03 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr21_+_40092301 | 0.03 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr9_-_13355071 | 0.03 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr3_+_32862730 | 0.03 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr7_+_50109239 | 0.03 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pdx1_pbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.3 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.0 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.0 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.0 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.2 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) anterograde axonal transport of mitochondrion(GO:0098957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |