Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
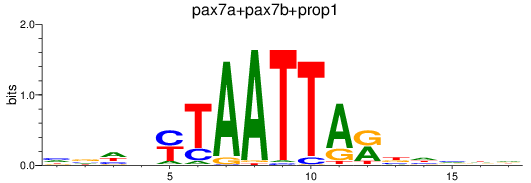
Results for pax7a+pax7b+prop1
Z-value: 0.54
Transcription factors associated with pax7a+pax7b+prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prop1
|
ENSDARG00000039756 | PROP paired-like homeobox 1 |
|
pax7b
|
ENSDARG00000070818 | paired box 7b |
|
pax7a
|
ENSDARG00000100398 | paired box 7a |
|
prop1
|
ENSDARG00000109950 | PROP paired-like homeobox 1 |
|
pax7a
|
ENSDARG00000111233 | paired box 7a |
|
prop1
|
ENSDARG00000114449 | PROP paired-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax7a | dr11_v1_chr11_+_41242644_41242644 | -0.55 | 1.7e-02 | Click! |
| prop1 | dr11_v1_chr13_+_8677166_8677304 | -0.49 | 4.0e-02 | Click! |
| pax7b | dr11_v1_chr23_+_20863145_20863145 | -0.19 | 4.6e-01 | Click! |
Activity profile of pax7a+pax7b+prop1 motif
Sorted Z-values of pax7a+pax7b+prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_6253246 | 1.39 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr20_+_48116476 | 1.29 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr20_+_54304800 | 1.16 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr10_+_15255198 | 1.13 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr9_+_8396755 | 1.03 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr9_+_8390737 | 0.99 |
ENSDART00000190891
ENSDART00000176877 ENSDART00000144851 ENSDART00000133880 ENSDART00000142233 |
zgc:113984
|
zgc:113984 |
| chr24_-_38110779 | 0.96 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr15_+_34934568 | 0.90 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr22_+_11972786 | 0.81 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr10_-_34002185 | 0.72 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr21_+_31150438 | 0.72 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr14_-_14659023 | 0.68 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr15_-_46779934 | 0.65 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr2_+_8112449 | 0.61 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr23_-_14766902 | 0.61 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr8_+_45334255 | 0.60 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr15_+_34592215 | 0.57 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr5_-_68333081 | 0.57 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr13_-_8692860 | 0.56 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr10_-_34916208 | 0.55 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr21_-_2209714 | 0.53 |
ENSDART00000163659
|
zgc:162971
|
zgc:162971 |
| chr12_-_48188928 | 0.51 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr12_-_33357655 | 0.51 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_31150773 | 0.50 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr6_-_15491579 | 0.49 |
ENSDART00000156439
|
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr17_-_4245902 | 0.49 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr5_-_3927692 | 0.49 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr24_-_11908115 | 0.47 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr19_+_19976990 | 0.46 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr12_+_22580579 | 0.45 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr25_-_37314700 | 0.45 |
ENSDART00000017807
|
igl4v8
|
immunoglobulin light 4 variable 8 |
| chr6_+_21001264 | 0.45 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr19_+_45962016 | 0.44 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr10_-_45058886 | 0.44 |
ENSDART00000159347
|
mrps24
|
mitochondrial ribosomal protein S24 |
| chr25_+_36292057 | 0.44 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr20_+_54274431 | 0.44 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr13_-_38039871 | 0.43 |
ENSDART00000140645
|
CR456624.1
|
|
| chr25_-_27621268 | 0.43 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr6_-_15492030 | 0.43 |
ENSDART00000156141
ENSDART00000183992 |
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr5_+_37903790 | 0.43 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr17_-_40956035 | 0.42 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr16_+_47207691 | 0.42 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr19_-_26923273 | 0.41 |
ENSDART00000089609
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr22_-_26865361 | 0.41 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr2_+_8780443 | 0.41 |
ENSDART00000137768
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr4_-_23759192 | 0.41 |
ENSDART00000014685
ENSDART00000131690 |
cpm
|
carboxypeptidase M |
| chr17_+_51682429 | 0.41 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr8_+_21159122 | 0.40 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr17_-_13007484 | 0.40 |
ENSDART00000156812
|
si:dkeyp-33b5.4
|
si:dkeyp-33b5.4 |
| chr21_-_39177564 | 0.40 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr20_-_37813863 | 0.40 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_+_9873750 | 0.39 |
ENSDART00000173348
ENSDART00000172813 |
lins1
|
lines homolog 1 |
| chr15_+_29727799 | 0.39 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr22_-_19102256 | 0.38 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr16_-_41646164 | 0.38 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr17_+_24821627 | 0.38 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr11_+_17984354 | 0.37 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr1_-_56948694 | 0.37 |
ENSDART00000152597
|
si:ch211-1f22.1
|
si:ch211-1f22.1 |
| chr11_-_41220794 | 0.37 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr4_+_4849789 | 0.37 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr2_-_9818640 | 0.36 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr9_+_54039006 | 0.36 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr13_-_9841806 | 0.36 |
ENSDART00000101949
|
sfxn4
|
sideroflexin 4 |
| chr7_+_51774502 | 0.35 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr18_-_26715655 | 0.35 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr21_+_15592426 | 0.35 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr2_-_42173834 | 0.35 |
ENSDART00000098357
ENSDART00000144707 |
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr17_+_12942634 | 0.35 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr23_-_19225709 | 0.35 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr4_-_77218637 | 0.34 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr9_-_43644261 | 0.34 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr8_+_3426522 | 0.33 |
ENSDART00000159466
|
CU929199.1
|
|
| chr25_+_22320738 | 0.33 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr11_+_17984167 | 0.32 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr5_-_20491198 | 0.32 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr15_-_2519640 | 0.32 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr9_-_50000144 | 0.31 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr4_-_73756673 | 0.31 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr21_+_27513859 | 0.31 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr25_+_37293312 | 0.31 |
ENSDART00000086737
ENSDART00000161595 |
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr11_-_40204621 | 0.31 |
ENSDART00000086287
|
znf362a
|
zinc finger protein 362a |
| chr8_-_25336589 | 0.30 |
ENSDART00000009682
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr9_+_18829360 | 0.30 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr20_-_26822522 | 0.30 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr3_+_40164129 | 0.30 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr1_-_55068941 | 0.29 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr7_+_24023653 | 0.29 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr6_-_7061086 | 0.29 |
ENSDART00000122719
|
ihhb
|
Indian hedgehog homolog b |
| chr1_+_35985813 | 0.29 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr8_+_20455134 | 0.29 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr8_+_32747612 | 0.28 |
ENSDART00000142824
|
hmcn2
|
hemicentin 2 |
| chr4_-_2637689 | 0.28 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr4_-_4834347 | 0.28 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr13_+_35474235 | 0.27 |
ENSDART00000181927
|
mkks
|
McKusick-Kaufman syndrome |
| chr22_-_10541372 | 0.27 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr9_+_23895711 | 0.27 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr20_-_7583486 | 0.27 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr21_-_32060993 | 0.26 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr19_+_2835240 | 0.26 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr8_+_26059677 | 0.26 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr25_+_16116740 | 0.26 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr6_-_55399214 | 0.26 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr5_+_28497956 | 0.26 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr2_+_50608099 | 0.25 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr1_+_12295142 | 0.25 |
ENSDART00000158595
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr18_+_19456648 | 0.25 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr5_+_56277866 | 0.25 |
ENSDART00000170610
ENSDART00000028854 ENSDART00000148749 |
aatf
|
apoptosis antagonizing transcription factor |
| chr12_-_47648538 | 0.25 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr10_+_42521943 | 0.25 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr18_+_17725410 | 0.25 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr19_-_18127808 | 0.24 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr1_+_33697170 | 0.24 |
ENSDART00000131664
|
NSUN3
|
NOP2/Sun RNA methyltransferase family member 3 |
| chr22_+_737211 | 0.24 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr5_-_5326010 | 0.24 |
ENSDART00000161946
|
pbx3a
|
pre-B-cell leukemia homeobox 3a |
| chr23_+_26026383 | 0.24 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr20_-_52902693 | 0.23 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr17_+_38030327 | 0.23 |
ENSDART00000085481
|
slc25a21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr24_-_25166720 | 0.23 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr5_+_60590796 | 0.23 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr23_+_36460239 | 0.23 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr18_-_1258777 | 0.23 |
ENSDART00000077106
ENSDART00000129065 |
ugt5f1
|
UDP glucuronosyltransferase 5 family, polypeptide F1 |
| chr6_-_35052145 | 0.23 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr9_-_9415000 | 0.22 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr23_-_41965557 | 0.22 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr24_-_36238054 | 0.22 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr10_+_45089820 | 0.22 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr6_+_7533601 | 0.22 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr6_+_40922572 | 0.22 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr13_+_8693410 | 0.21 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr21_+_15603597 | 0.21 |
ENSDART00000138626
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr16_+_23303859 | 0.21 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr8_+_31717175 | 0.21 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr25_-_20024829 | 0.20 |
ENSDART00000140182
ENSDART00000174776 |
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr6_+_19689464 | 0.20 |
ENSDART00000164960
|
stx8
|
syntaxin 8 |
| chr24_-_24724233 | 0.20 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr8_+_11325310 | 0.20 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr8_+_18011522 | 0.20 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr9_-_11676491 | 0.19 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr9_-_27398369 | 0.19 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr20_-_54198130 | 0.19 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr19_+_5480327 | 0.19 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr10_+_45071603 | 0.19 |
ENSDART00000186505
ENSDART00000157573 |
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr6_+_40951227 | 0.19 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr2_+_24374669 | 0.19 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr24_+_21540842 | 0.19 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr17_-_16422654 | 0.19 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr15_-_16155729 | 0.18 |
ENSDART00000192212
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr14_-_26425416 | 0.18 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr20_+_19006703 | 0.18 |
ENSDART00000128435
|
pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr11_-_41853874 | 0.18 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr22_-_20695237 | 0.18 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr16_-_43344859 | 0.18 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr7_-_69185124 | 0.18 |
ENSDART00000182217
ENSDART00000191359 |
usp10
|
ubiquitin specific peptidase 10 |
| chr19_-_425145 | 0.18 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr19_-_5103141 | 0.18 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_-_22242884 | 0.18 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr7_+_20383841 | 0.18 |
ENSDART00000052906
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr3_-_15667713 | 0.18 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr24_+_21514283 | 0.18 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr8_+_11425048 | 0.17 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr9_+_34127005 | 0.17 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr25_-_6292560 | 0.17 |
ENSDART00000153496
|
wdr61
|
WD repeat domain 61 |
| chr8_+_18555559 | 0.17 |
ENSDART00000149523
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr5_-_1203455 | 0.17 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr3_-_26787430 | 0.17 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr4_+_5249494 | 0.17 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr8_-_37249813 | 0.16 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr11_+_1512571 | 0.16 |
ENSDART00000027386
|
acot8
|
acyl-CoA thioesterase 8 |
| chr18_-_45761868 | 0.16 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr13_-_9213207 | 0.16 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr18_+_45958375 | 0.16 |
ENSDART00000108636
|
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr1_+_51721851 | 0.16 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr25_-_32888115 | 0.16 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr7_+_24573721 | 0.16 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr13_+_2442841 | 0.15 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr18_-_29896367 | 0.15 |
ENSDART00000191303
|
cmc2
|
C-x(9)-C motif containing 2 |
| chr23_-_29394505 | 0.15 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr18_+_14619544 | 0.15 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr9_-_43538328 | 0.15 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr10_+_36092564 | 0.15 |
ENSDART00000086490
|
smyd4
|
SET and MYND domain containing 4 |
| chr9_+_48761455 | 0.15 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr15_-_17618800 | 0.15 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr18_-_19456269 | 0.15 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr7_+_5956937 | 0.14 |
ENSDART00000170763
|
hist1h2a4
|
histone cluster 1 H2A family member 4 |
| chr24_+_40860320 | 0.14 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr6_-_35052388 | 0.14 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr24_+_21621654 | 0.14 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr10_-_43721530 | 0.13 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr1_+_49668423 | 0.13 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr23_-_33679579 | 0.13 |
ENSDART00000188674
|
tfcp2
|
transcription factor CP2 |
| chr8_-_15129573 | 0.13 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr6_+_13024448 | 0.13 |
ENSDART00000104768
|
pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr10_+_11261576 | 0.13 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr10_+_45059702 | 0.13 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr17_+_28533102 | 0.13 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr7_+_27898091 | 0.12 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr22_-_10541712 | 0.12 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr23_+_42272588 | 0.12 |
ENSDART00000164907
|
CABZ01065131.1
|
|
| chr1_-_15797663 | 0.12 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr2_-_57264262 | 0.12 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax7a+pax7b+prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 1.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.7 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.5 | GO:0060986 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.4 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.4 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.4 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.8 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.3 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.6 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.3 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.3 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.4 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0030183 | B cell differentiation(GO:0030183) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.6 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.6 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 1.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.3 | 2.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 1.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |