Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
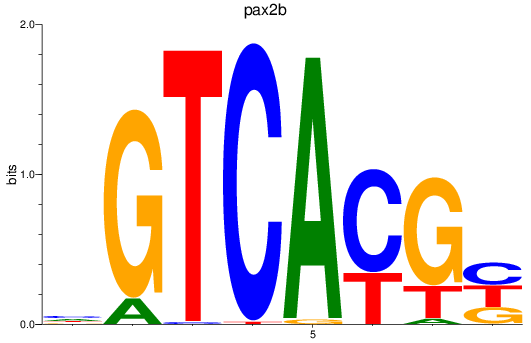
Results for pax2b
Z-value: 2.00
Transcription factors associated with pax2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2b
|
ENSDARG00000032578 | paired box 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax2b | dr11_v1_chr12_-_45876387_45876387 | -0.84 | 1.2e-05 | Click! |
Activity profile of pax2b motif
Sorted Z-values of pax2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_33359052 | 6.78 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr22_-_25612680 | 6.72 |
ENSDART00000114167
|
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr21_-_19919918 | 5.43 |
ENSDART00000137307
ENSDART00000142523 ENSDART00000065670 |
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr3_-_32362872 | 5.39 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr8_-_22542467 | 4.90 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr23_-_22523303 | 4.88 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr11_+_45153104 | 4.77 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr2_+_12255568 | 4.75 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr1_+_45707219 | 4.70 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr8_+_52442622 | 4.66 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr7_+_27455321 | 4.49 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr7_+_67699009 | 4.34 |
ENSDART00000192810
|
zgc:162592
|
zgc:162592 |
| chr22_-_17677947 | 4.30 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr16_+_30117798 | 4.27 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr8_+_52442785 | 4.20 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr8_+_11471350 | 4.18 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr19_-_27827744 | 4.10 |
ENSDART00000181620
|
papd7
|
PAP associated domain containing 7 |
| chr6_-_53143667 | 4.10 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr10_+_36662640 | 4.07 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr15_-_35126332 | 3.89 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr4_+_23117557 | 3.86 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr1_+_6135176 | 3.84 |
ENSDART00000092324
ENSDART00000179970 |
abcb6a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6a |
| chr11_+_13041341 | 3.82 |
ENSDART00000166126
ENSDART00000162074 ENSDART00000170889 |
btf3l4
|
basic transcription factor 3-like 4 |
| chr8_+_16758304 | 3.82 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr11_-_44801968 | 3.77 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr19_+_791538 | 3.75 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr7_+_41812190 | 3.74 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr19_-_874888 | 3.71 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr2_+_42871831 | 3.68 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr14_+_8947282 | 3.65 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr11_+_13176568 | 3.63 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr15_-_35112937 | 3.52 |
ENSDART00000154565
ENSDART00000099642 |
zgc:77118
|
zgc:77118 |
| chr19_-_47570672 | 3.49 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr22_+_336256 | 3.41 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr16_+_10557504 | 3.38 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr14_-_16810401 | 3.36 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr3_+_40576447 | 3.30 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr13_-_45022527 | 3.27 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr6_-_12135741 | 3.24 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr2_+_24936766 | 3.23 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr17_+_25187226 | 3.22 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr19_+_15441022 | 3.17 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_+_15048410 | 3.11 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr10_+_23060391 | 3.08 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr6_+_153146 | 3.02 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr21_+_7605803 | 2.99 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr23_+_7692042 | 2.98 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr10_-_24765988 | 2.96 |
ENSDART00000064463
|
timm10b
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr12_-_25887864 | 2.94 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr14_+_35414632 | 2.90 |
ENSDART00000191516
ENSDART00000084914 |
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr22_+_25184459 | 2.84 |
ENSDART00000105308
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr5_+_44846280 | 2.83 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr5_-_37047147 | 2.82 |
ENSDART00000097712
ENSDART00000158974 |
ftr99
|
finTRIM family, member 99 |
| chr11_+_24758967 | 2.82 |
ENSDART00000005616
ENSDART00000133481 |
rnpep
|
arginyl aminopeptidase (aminopeptidase B) |
| chr5_+_44846434 | 2.82 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr3_-_15475067 | 2.75 |
ENSDART00000025324
ENSDART00000139575 |
spns1
|
spinster homolog 1 (Drosophila) |
| chr7_-_51773166 | 2.74 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr16_+_20496691 | 2.74 |
ENSDART00000182737
ENSDART00000078984 |
cpvl
|
carboxypeptidase, vitellogenic-like |
| chr19_+_31585341 | 2.74 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr19_+_15440841 | 2.71 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr3_-_26191960 | 2.70 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr7_+_51795667 | 2.70 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr9_-_27868267 | 2.70 |
ENSDART00000079502
|
dbr1
|
debranching RNA lariats 1 |
| chr17_+_25833947 | 2.68 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr15_+_29024895 | 2.68 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr20_-_40367493 | 2.67 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr17_+_28005763 | 2.62 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr2_+_41526904 | 2.60 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr22_+_25086942 | 2.60 |
ENSDART00000061117
|
rrbp1b
|
ribosome binding protein 1b |
| chr7_+_47243564 | 2.57 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr20_-_36671660 | 2.56 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr4_-_9780931 | 2.55 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr5_-_69944084 | 2.55 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr2_+_32743807 | 2.55 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr14_+_20156477 | 2.54 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr6_-_26080384 | 2.52 |
ENSDART00000157181
ENSDART00000154568 |
hs2st1b
|
heparan sulfate 2-O-sulfotransferase 1b |
| chr9_+_426392 | 2.50 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr17_+_20569806 | 2.47 |
ENSDART00000113936
|
SAMD8
|
zgc:162183 |
| chr3_-_37785873 | 2.46 |
ENSDART00000011691
|
baxa
|
BCL2 associated X, apoptosis regulator a |
| chr14_-_33277743 | 2.46 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr8_+_12951155 | 2.45 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr5_+_4533244 | 2.44 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr14_+_24840669 | 2.44 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr5_+_33301005 | 2.43 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr23_+_33963619 | 2.43 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr9_-_12269847 | 2.41 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr23_+_40139765 | 2.41 |
ENSDART00000185376
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr4_+_25912308 | 2.40 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr16_+_28547157 | 2.39 |
ENSDART00000109450
ENSDART00000165687 |
fam171a1
|
family with sequence similarity 171, member A1 |
| chr22_+_25086567 | 2.39 |
ENSDART00000192114
ENSDART00000177284 ENSDART00000180296 ENSDART00000190384 |
rrbp1b
|
ribosome binding protein 1b |
| chr5_-_1999417 | 2.37 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr24_-_21903588 | 2.35 |
ENSDART00000180991
|
spata13
|
spermatogenesis associated 13 |
| chr24_-_21903360 | 2.34 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr5_+_52596850 | 2.34 |
ENSDART00000162022
|
ptar1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr15_+_45595385 | 2.33 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr6_+_4255319 | 2.33 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr21_+_21743599 | 2.33 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr12_-_17686404 | 2.32 |
ENSDART00000079065
|
ccz1
|
CCZ1 homolog, vacuolar protein trafficking and biogenesis associated |
| chr4_+_25912654 | 2.32 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr9_-_41090048 | 2.32 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr13_+_25505580 | 2.32 |
ENSDART00000140634
|
inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr6_-_13716744 | 2.32 |
ENSDART00000111102
|
chpfb
|
chondroitin polymerizing factor b |
| chr13_-_8304605 | 2.32 |
ENSDART00000080460
|
atl2
|
atlastin GTPase 2 |
| chr5_+_41476443 | 2.31 |
ENSDART00000145228
ENSDART00000137981 ENSDART00000142538 |
pias2
|
protein inhibitor of activated STAT, 2 |
| chr7_-_44605050 | 2.31 |
ENSDART00000148471
ENSDART00000149072 |
tk2
|
thymidine kinase 2, mitochondrial |
| chr7_-_42206720 | 2.30 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr5_-_22130937 | 2.30 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr13_-_45022301 | 2.27 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr8_+_13700605 | 2.27 |
ENSDART00000144516
|
lonrf1l
|
LON peptidase N-terminal domain and ring finger 1, like |
| chr13_-_6250317 | 2.26 |
ENSDART00000180416
|
tuba4l
|
tubulin, alpha 4 like |
| chr21_-_34972872 | 2.26 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr14_+_30413312 | 2.25 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr3_+_24094581 | 2.24 |
ENSDART00000138270
ENSDART00000131509 |
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr1_+_19764995 | 2.24 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr7_+_66634167 | 2.21 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr1_-_14506759 | 2.20 |
ENSDART00000057044
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr20_+_28364742 | 2.20 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr14_-_33478963 | 2.19 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr9_-_19366538 | 2.16 |
ENSDART00000138431
|
zgc:152951
|
zgc:152951 |
| chr6_+_20954400 | 2.15 |
ENSDART00000143248
ENSDART00000165806 |
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr2_-_4787566 | 2.14 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr13_-_12389748 | 2.14 |
ENSDART00000141606
|
commd8
|
COMM domain containing 8 |
| chr15_-_20412286 | 2.13 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr6_-_41138854 | 2.11 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr11_+_41838801 | 2.10 |
ENSDART00000014871
|
akr7a3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr3_+_19245804 | 2.10 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr12_+_19408373 | 2.10 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr8_+_50190742 | 2.09 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr24_-_17023392 | 2.08 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr15_-_37104165 | 2.08 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr11_-_34480822 | 2.07 |
ENSDART00000129029
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr20_-_40766387 | 2.06 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr13_+_2357637 | 2.06 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr6_+_3717613 | 2.05 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr15_-_41311292 | 2.03 |
ENSDART00000133835
ENSDART00000085645 ENSDART00000134075 |
usp16
|
ubiquitin specific peptidase 16 |
| chr12_-_33770299 | 2.03 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr14_+_164556 | 2.03 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr6_+_3716666 | 2.01 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr11_+_5499661 | 2.01 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr3_-_13546610 | 2.01 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr9_-_14273652 | 2.01 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr24_+_33462800 | 2.00 |
ENSDART00000166666
ENSDART00000050826 |
rmc1
|
regulator of MON1-CCZ1 |
| chr15_+_1534644 | 2.00 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr25_-_13871118 | 1.99 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr8_+_28724692 | 1.99 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr10_-_7702029 | 1.99 |
ENSDART00000018017
|
gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr15_+_46853252 | 1.98 |
ENSDART00000186040
|
zgc:153039
|
zgc:153039 |
| chr15_+_29025090 | 1.97 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr22_-_21676364 | 1.97 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr12_+_16953415 | 1.96 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr16_+_43077909 | 1.95 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr1_+_47499888 | 1.95 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr15_-_33304133 | 1.95 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr7_-_41013575 | 1.93 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr13_+_21676235 | 1.93 |
ENSDART00000137804
ENSDART00000134950 ENSDART00000129653 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr1_+_5422439 | 1.92 |
ENSDART00000055047
ENSDART00000142248 |
stk16
|
serine/threonine kinase 16 |
| chr13_-_50463938 | 1.91 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr16_-_7793457 | 1.91 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr8_-_410199 | 1.91 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr23_-_21763598 | 1.90 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr7_+_35193832 | 1.90 |
ENSDART00000189002
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr10_-_24724388 | 1.88 |
ENSDART00000148582
|
smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr12_+_19958845 | 1.87 |
ENSDART00000193248
|
ercc4
|
excision repair cross-complementation group 4 |
| chr8_-_410728 | 1.86 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr5_-_24029228 | 1.86 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr24_-_6024466 | 1.86 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr23_+_24973773 | 1.85 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr13_+_36595618 | 1.85 |
ENSDART00000022684
|
cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr16_-_8120203 | 1.85 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr13_+_421231 | 1.84 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr16_+_26732086 | 1.83 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr22_+_30047245 | 1.83 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr25_-_6049339 | 1.83 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr11_-_43226255 | 1.82 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr8_+_21159122 | 1.82 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr4_+_76304911 | 1.81 |
ENSDART00000172734
ENSDART00000161850 |
si:ch73-389k6.1
|
si:ch73-389k6.1 |
| chr10_-_41352502 | 1.81 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr13_-_18011168 | 1.81 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr8_+_48966165 | 1.81 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr22_-_11517377 | 1.80 |
ENSDART00000193885
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr5_-_67365333 | 1.78 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr14_+_1170968 | 1.77 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr14_+_46118834 | 1.77 |
ENSDART00000124417
ENSDART00000017785 |
naa15a
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit a |
| chr14_+_16034447 | 1.77 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr7_-_38570299 | 1.77 |
ENSDART00000143815
|
celf1
|
cugbp, Elav-like family member 1 |
| chr25_-_14424406 | 1.77 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr20_+_2642855 | 1.76 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr21_-_25295087 | 1.76 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr7_-_55648336 | 1.76 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr19_-_46088429 | 1.76 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr3_-_40976288 | 1.75 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr24_-_38192003 | 1.75 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr2_+_23081247 | 1.75 |
ENSDART00000099702
ENSDART00000088867 |
mfsd12a
|
major facilitator superfamily domain containing 12a |
| chr7_-_32895668 | 1.74 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr13_+_46803979 | 1.74 |
ENSDART00000159260
|
CU695232.1
|
|
| chr23_-_270847 | 1.73 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr25_-_2723902 | 1.73 |
ENSDART00000143721
ENSDART00000175224 |
adpgk
|
ADP-dependent glucokinase |
| chr5_+_37343643 | 1.72 |
ENSDART00000163993
ENSDART00000164459 ENSDART00000167006 ENSDART00000128122 ENSDART00000183720 |
klhl13
|
kelch-like family member 13 |
| chr7_+_42206847 | 1.72 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr5_-_31689796 | 1.71 |
ENSDART00000184319
ENSDART00000190229 ENSDART00000186294 ENSDART00000170313 ENSDART00000147065 ENSDART00000134427 ENSDART00000098172 |
sh3glb2b
|
SH3-domain GRB2-like endophilin B2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 1.6 | 4.8 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.3 | 5.4 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 1.2 | 3.7 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.9 | 1.9 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.8 | 3.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.8 | 2.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.8 | 3.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.7 | 4.5 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.7 | 2.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.7 | 3.6 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.7 | 3.5 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.7 | 3.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.7 | 2.7 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.7 | 2.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.7 | 3.4 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.7 | 3.3 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.7 | 2.6 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.6 | 1.9 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.6 | 2.6 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.6 | 1.9 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.6 | 3.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.6 | 1.8 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.6 | 1.7 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.6 | 1.7 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.5 | 2.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.5 | 1.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.5 | 4.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.5 | 2.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.5 | 1.5 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.5 | 1.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.5 | 2.3 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.5 | 4.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.5 | 1.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.4 | 2.7 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.4 | 1.8 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.4 | 4.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.4 | 1.3 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.4 | 2.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.4 | 8.3 | GO:0032885 | regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.4 | 1.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.4 | 1.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.4 | 2.0 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 1.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.4 | 4.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 1.1 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.4 | 1.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.4 | 1.8 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.4 | 1.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 1.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 3.1 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.3 | 1.6 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 1.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 3.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 1.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.3 | 3.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 2.7 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.3 | 1.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.3 | 1.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.3 | 1.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 2.9 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.3 | 6.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 0.9 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.3 | 1.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 0.8 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.3 | 2.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 2.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 1.4 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.3 | 3.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 1.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.0 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 1.3 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.3 | 1.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 2.0 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.2 | 1.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 2.5 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.2 | 2.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.9 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 0.9 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 2.0 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 1.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 1.7 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.2 | 4.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 1.0 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 2.4 | GO:0006999 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.2 | 1.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.0 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.2 | 1.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.9 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 1.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 3.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 5.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.7 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 3.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 0.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 1.1 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.2 | 0.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.9 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 0.5 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.2 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.2 | 1.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.0 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 3.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 4.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 1.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.6 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 2.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.5 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 7.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 3.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 2.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 2.7 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 2.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.9 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.9 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 2.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.9 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 5.2 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 0.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 2.9 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.1 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 2.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 3.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 3.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.8 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 2.0 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.5 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 1.3 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.7 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 2.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.9 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 1.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 3.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 2.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.5 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) heterochromatin organization(GO:0070828) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 3.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.8 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.7 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 7.6 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 0.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 2.0 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 0.5 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 2.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.5 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 1.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 1.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.7 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.3 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.5 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 2.4 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 0.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.5 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 10.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.4 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.2 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 2.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 3.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 2.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.0 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 3.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.8 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 1.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 1.5 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 2.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 2.3 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 1.1 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.7 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 1.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.5 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 4.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.2 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.2 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.9 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 2.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.2 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 | 1.3 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 2.6 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 1.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 3.1 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 1.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.1 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.7 | GO:0048599 | oocyte differentiation(GO:0009994) oocyte development(GO:0048599) |
| 0.0 | 2.6 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.0 | GO:0002688 | regulation of leukocyte chemotaxis(GO:0002688) |
| 0.0 | 2.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 2.9 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.5 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 1.1 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0006620 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) posttranslational protein targeting to membrane(GO:0006620) posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 1.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.1 | GO:0043102 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.5 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.7 | 12.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.7 | 2.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.6 | 1.9 | GO:1990879 | CST complex(GO:1990879) |
| 0.6 | 1.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.6 | 2.5 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.5 | 2.6 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.5 | 1.5 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.5 | 1.9 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.5 | 2.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 2.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 2.4 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.4 | 1.5 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.4 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.3 | 1.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 1.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 3.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 2.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 1.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 0.8 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.3 | 1.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 2.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 3.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 1.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 2.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 4.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.6 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.2 | 1.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 2.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 1.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 0.5 | GO:0072380 | TRC complex(GO:0072380) |
| 0.2 | 0.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 0.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 5.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.4 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 5.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 15.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 13.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.7 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.4 | GO:0030670 | endocytic vesicle membrane(GO:0030666) phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.5 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 2.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.4 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 4.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.5 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 3.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 3.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 5.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.8 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 4.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 9.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 2.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 3.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 2.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 9.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.0 | 4.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.0 | 7.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.9 | 4.4 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.8 | 2.5 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.8 | 12.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.8 | 3.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.8 | 2.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.7 | 5.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.6 | 2.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.6 | 3.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.6 | 3.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 1.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.6 | 6.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 2.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.5 | 4.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 2.7 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.5 | 1.5 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 3.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.5 | 2.0 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.5 | 1.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.5 | 1.5 | GO:0034618 | arginine binding(GO:0034618) |
| 0.5 | 5.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 2.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 4.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 1.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 4.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.4 | 5.4 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.4 | 1.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.4 | 1.5 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.4 | 1.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.4 | 1.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 1.0 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.3 | 1.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 1.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 2.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 4.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 1.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 2.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.3 | 1.5 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.3 | 2.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.3 | 2.9 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.3 | 0.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 6.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 0.9 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.3 | 0.8 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.3 | 2.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.3 | 1.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.7 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 1.5 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.2 | 1.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 1.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 1.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 4.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 3.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 0.7 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 0.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 2.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.1 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 1.5 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.2 | 2.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 1.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 2.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.7 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.0 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 1.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 1.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 0.7 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 1.9 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.2 | 3.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 1.4 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.2 | 4.2 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 0.7 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.2 | 2.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 3.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 0.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.2 | 1.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.4 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 1.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 4.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 3.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 6.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.5 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 1.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.1 | 0.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.9 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 2.7 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 3.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 3.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 6.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.1 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.9 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase activity(GO:0004448) isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 2.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 3.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.3 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 4.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.2 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.1 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.9 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 2.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 2.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 11.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 3.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 6.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 1.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 13.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 3.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.4 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 4.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 4.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 6.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 4.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.3 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 2.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 3.0 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 7.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 8.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 10.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 6.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 2.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 6.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 1.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 2.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 4.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 1.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 3.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 3.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 2.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 3.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 0.8 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.2 | 1.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 3.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 2.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 2.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.0 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 4.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.6 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 1.9 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE EXCISION REPAIR | Genes involved in Nucleotide Excision Repair |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.7 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |