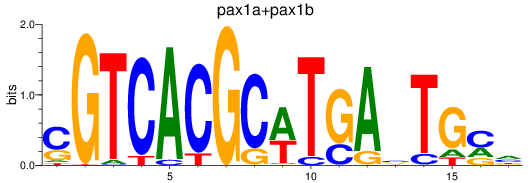
Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for pax1a+pax1b_pax9
Z-value: 0.22
Transcription factors associated with pax1a+pax1b_pax9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax1a
|
ENSDARG00000008203 | paired box 1a |
|
pax1b
|
ENSDARG00000073814 | paired box 1b |
|
pax9
|
ENSDARG00000053829 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax1b | dr11_v1_chr20_-_48701593_48701593 | -0.82 | 3.5e-05 | Click! |
| pax9 | dr11_v1_chr17_-_38244117_38244117 | 0.78 | 1.2e-04 | Click! |
| pax1a | dr11_v1_chr17_+_42274240_42274240 | 0.63 | 5.5e-03 | Click! |
Activity profile of pax1a+pax1b_pax9 motif
Sorted Z-values of pax1a+pax1b_pax9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_956821 | 0.50 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr18_-_48550426 | 0.44 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr10_-_39154778 | 0.41 |
ENSDART00000186811
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr2_+_49864219 | 0.38 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr21_+_13861589 | 0.33 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr24_+_41690545 | 0.31 |
ENSDART00000160069
|
lama1
|
laminin, alpha 1 |
| chr17_+_996509 | 0.30 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr20_-_31075972 | 0.29 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr12_-_7854216 | 0.29 |
ENSDART00000149594
|
ank3b
|
ankyrin 3b |
| chr5_-_35743741 | 0.23 |
ENSDART00000046157
ENSDART00000123562 |
ophn1
|
oligophrenin 1 |
| chr14_+_23518110 | 0.23 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr2_+_19633493 | 0.21 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr1_-_46343999 | 0.15 |
ENSDART00000145117
ENSDART00000193233 |
atp11a
|
ATPase phospholipid transporting 11A |
| chr2_-_44282796 | 0.14 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr6_-_41079209 | 0.13 |
ENSDART00000151592
|
rab44
|
RAB44, member RAS oncogene family |
| chr3_-_50277959 | 0.12 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr1_+_12195700 | 0.12 |
ENSDART00000040307
|
tdrd7a
|
tudor domain containing 7 a |
| chr12_-_18483348 | 0.11 |
ENSDART00000152757
|
tex2l
|
testis expressed 2, like |
| chr11_-_165288 | 0.11 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr2_-_44283554 | 0.10 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr20_+_12702923 | 0.10 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr3_+_47322494 | 0.09 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr5_+_39875646 | 0.09 |
ENSDART00000097608
|
si:ch73-138i16.1
|
si:ch73-138i16.1 |
| chr11_+_37278457 | 0.09 |
ENSDART00000188946
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr6_-_31224563 | 0.08 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr1_+_39553040 | 0.05 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr5_-_1047504 | 0.05 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr10_-_39154594 | 0.04 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr16_+_20738740 | 0.03 |
ENSDART00000079343
|
jazf1b
|
JAZF zinc finger 1b |
| chr20_+_29587995 | 0.02 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr5_+_39936896 | 0.02 |
ENSDART00000135126
|
si:ch73-138i16.1
|
si:ch73-138i16.1 |
| chr23_+_43255328 | 0.02 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr13_-_24379271 | 0.01 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr25_+_6306885 | 0.01 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr9_-_24031461 | 0.01 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr4_-_8902406 | 0.01 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr2_+_394166 | 0.00 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax1a+pax1b_pax9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |