Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
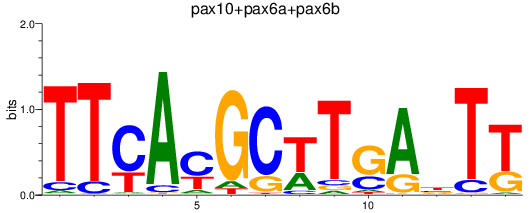
Results for pax10+pax6a+pax6b
Z-value: 1.31
Transcription factors associated with pax10+pax6a+pax6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax6b
|
ENSDARG00000045936 | paired box 6b |
|
pax10
|
ENSDARG00000053364 | paired box 10 |
|
pax6a
|
ENSDARG00000103379 | paired box 6a |
|
pax10
|
ENSDARG00000111379 | paired box 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax6a | dr11_v1_chr25_-_15049694_15049781 | -0.99 | 4.5e-14 | Click! |
| pax10 | dr11_v1_chr3_+_32553714_32553714 | -0.64 | 4.3e-03 | Click! |
| pax6b | dr11_v1_chr7_+_15872357_15872357 | -0.48 | 4.5e-02 | Click! |
Activity profile of pax10+pax6a+pax6b motif
Sorted Z-values of pax10+pax6a+pax6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_8046764 | 6.31 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr10_-_8053385 | 5.73 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr25_+_22320738 | 5.19 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr3_+_43086548 | 4.99 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr8_+_52442622 | 4.82 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr10_-_8053753 | 4.44 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr8_+_52442785 | 4.35 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr2_+_6255434 | 3.28 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr5_+_57924611 | 3.09 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr21_-_2322102 | 2.94 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr16_+_39159752 | 2.89 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr7_-_20582842 | 2.81 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr9_+_23003208 | 2.78 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr19_-_42503143 | 2.73 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr9_+_8365398 | 2.73 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr3_-_26184018 | 2.56 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr23_+_17926279 | 2.51 |
ENSDART00000012540
|
chia.4
|
chitinase, acidic.4 |
| chr25_+_7532811 | 2.39 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr17_+_17804752 | 2.35 |
ENSDART00000123350
|
sptlc2a
|
serine palmitoyltransferase, long chain base subunit 2a |
| chr21_-_22117085 | 2.31 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr13_-_32726178 | 2.30 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr19_-_11015238 | 2.30 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr3_-_54607166 | 2.24 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr19_-_25114701 | 2.11 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr5_-_47975440 | 2.11 |
ENSDART00000145665
ENSDART00000007057 |
ccnh
|
cyclin H |
| chr23_-_27235403 | 1.91 |
ENSDART00000134418
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr4_-_13613148 | 1.88 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr3_+_25914064 | 1.87 |
ENSDART00000162558
ENSDART00000166843 |
tom1
|
target of myb1 membrane trafficking protein |
| chr13_-_25196758 | 1.87 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr1_+_2431956 | 1.86 |
ENSDART00000183832
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr1_-_29653472 | 1.82 |
ENSDART00000109224
|
si:dkey-1h24.2
|
si:dkey-1h24.2 |
| chr23_+_36460239 | 1.79 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr6_+_33076839 | 1.76 |
ENSDART00000073755
ENSDART00000122242 |
pomgnt1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr25_+_19870603 | 1.73 |
ENSDART00000047251
|
gramd4b
|
GRAM domain containing 4b |
| chr18_-_15269272 | 1.73 |
ENSDART00000048206
|
tmem263
|
transmembrane protein 263 |
| chr5_+_41477954 | 1.72 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr19_-_4793263 | 1.68 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr12_+_3571770 | 1.67 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr2_+_2772447 | 1.66 |
ENSDART00000124882
|
thoc1
|
THO complex 1 |
| chr5_+_60590796 | 1.62 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr23_-_24542156 | 1.61 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr16_-_25680666 | 1.60 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr15_-_1001177 | 1.56 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr5_-_26247973 | 1.55 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr20_-_3238110 | 1.55 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr11_-_44931962 | 1.51 |
ENSDART00000170345
|
pfklb
|
phosphofructokinase, liver b |
| chr18_+_14277003 | 1.49 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr21_-_32781612 | 1.45 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr2_+_25657958 | 1.43 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr7_+_26545911 | 1.43 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr5_+_41477526 | 1.39 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr4_-_4261673 | 1.38 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr12_-_48374728 | 1.36 |
ENSDART00000153403
ENSDART00000188117 |
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr12_+_23812530 | 1.33 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr23_-_27505825 | 1.32 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr12_+_47794089 | 1.31 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr10_-_14920989 | 1.31 |
ENSDART00000184617
|
smad2
|
SMAD family member 2 |
| chr15_+_39977461 | 1.29 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr23_-_15916316 | 1.29 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr7_-_58244220 | 1.29 |
ENSDART00000180450
|
unm_hu7910
|
un-named hu7910 |
| chr6_-_10034145 | 1.29 |
ENSDART00000185999
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr18_+_50924556 | 1.27 |
ENSDART00000159287
|
pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr2_+_11031360 | 1.22 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr9_+_43797902 | 1.20 |
ENSDART00000020550
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr23_-_18381361 | 1.20 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr24_+_11908833 | 1.19 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr19_-_3726768 | 1.19 |
ENSDART00000161738
|
smim13
|
small integral membrane protein 13 |
| chr5_-_55914268 | 1.18 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr23_+_25292147 | 1.18 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr25_+_7532627 | 1.18 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr24_-_41320037 | 1.16 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr9_-_50000144 | 1.14 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr9_-_746317 | 1.14 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr13_-_31164749 | 1.14 |
ENSDART00000049180
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr23_+_25291891 | 1.13 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr12_-_35582683 | 1.12 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr18_-_45761868 | 1.10 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr4_+_5341592 | 1.10 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr4_-_9196291 | 1.10 |
ENSDART00000153963
|
hcfc2
|
host cell factor C2 |
| chr24_-_2381143 | 1.10 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr23_+_44611864 | 1.09 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr5_+_62052750 | 1.08 |
ENSDART00000192103
ENSDART00000181866 |
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr24_+_11908480 | 1.08 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr24_+_26379441 | 1.08 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr21_-_7178348 | 1.05 |
ENSDART00000187467
|
fam69b
|
family with sequence similarity 69, member B |
| chr7_+_26545502 | 1.05 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr3_-_34717882 | 1.05 |
ENSDART00000151127
|
thraa
|
thyroid hormone receptor alpha a |
| chr4_+_76466751 | 1.05 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr9_-_31915423 | 1.05 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr18_-_21685055 | 1.04 |
ENSDART00000019861
|
mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr14_+_24934736 | 1.03 |
ENSDART00000191821
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr14_-_5407555 | 1.01 |
ENSDART00000001424
|
pcgf1
|
polycomb group ring finger 1 |
| chr15_-_704408 | 1.00 |
ENSDART00000156200
ENSDART00000166404 ENSDART00000131040 |
zgc:174574
|
zgc:174574 |
| chr3_-_23513155 | 0.99 |
ENSDART00000170200
|
BX682558.1
|
|
| chr5_+_40837539 | 0.98 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr4_+_13931578 | 0.98 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr4_-_13931508 | 0.96 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr10_-_167782 | 0.95 |
ENSDART00000108780
|
erg
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr11_-_11336986 | 0.95 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr2_+_21048661 | 0.94 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr18_-_20466061 | 0.93 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr23_+_44614056 | 0.91 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr7_-_30624435 | 0.89 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr13_-_31167461 | 0.89 |
ENSDART00000190640
ENSDART00000192002 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr4_-_13931293 | 0.89 |
ENSDART00000067172
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr11_+_19603251 | 0.86 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr7_-_45852270 | 0.86 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr17_-_41798856 | 0.86 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr3_+_60828813 | 0.85 |
ENSDART00000128260
|
CABZ01087514.1
|
|
| chr8_-_28357177 | 0.85 |
ENSDART00000182319
|
KLHL12 (1 of many)
|
kelch like family member 12 |
| chr20_+_27712714 | 0.84 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr10_+_24690534 | 0.84 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr23_+_30707837 | 0.83 |
ENSDART00000016096
|
dnajc11a
|
DnaJ (Hsp40) homolog, subfamily C, member 11a |
| chr15_+_1534644 | 0.83 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr14_-_14659023 | 0.82 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr5_-_33039670 | 0.82 |
ENSDART00000141361
|
glipr2
|
GLI pathogenesis-related 2 |
| chr25_-_10791437 | 0.81 |
ENSDART00000127054
|
BX572619.1
|
|
| chr14_-_45967981 | 0.80 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr5_+_38825225 | 0.80 |
ENSDART00000076835
|
mrpl1
|
mitochondrial ribosomal protein L1 |
| chr13_-_11378127 | 0.79 |
ENSDART00000158632
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr16_-_5105295 | 0.79 |
ENSDART00000082071
ENSDART00000148955 ENSDART00000184700 ENSDART00000188127 |
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr17_-_15029258 | 0.78 |
ENSDART00000168332
ENSDART00000012877 ENSDART00000109190 |
ero1a
|
endoplasmic reticulum oxidoreductase alpha |
| chr21_-_17016788 | 0.78 |
ENSDART00000186703
|
AL844518.1
|
|
| chr23_-_7125494 | 0.78 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr17_+_43926523 | 0.77 |
ENSDART00000121550
ENSDART00000041447 |
ktn1
|
kinectin 1 |
| chr12_+_33361948 | 0.77 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr24_+_37709191 | 0.76 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr11_+_45153104 | 0.75 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr23_-_2901167 | 0.75 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr7_-_19526721 | 0.74 |
ENSDART00000114203
|
man2b2
|
mannosidase, alpha, class 2B, member 2 |
| chr25_-_37284370 | 0.73 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr3_-_37351225 | 0.73 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr22_+_21398508 | 0.72 |
ENSDART00000089408
ENSDART00000186091 |
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr14_+_22172047 | 0.72 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr7_-_73843720 | 0.72 |
ENSDART00000111622
|
caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr2_-_48753873 | 0.72 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr12_-_35582521 | 0.71 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr11_-_4023940 | 0.71 |
ENSDART00000058728
ENSDART00000171245 |
nek4
|
NIMA-related kinase 4 |
| chr19_+_12413683 | 0.71 |
ENSDART00000151214
|
seh1l
|
SEH1-like (S. cerevisiae) |
| chr4_-_1818315 | 0.71 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr2_+_21000334 | 0.71 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr5_+_4054704 | 0.70 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr20_+_4334008 | 0.69 |
ENSDART00000168322
|
tiam2b
|
T cell lymphoma invasion and metastasis 2b |
| chr7_-_30174882 | 0.68 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr5_+_32162684 | 0.67 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr12_-_3077395 | 0.67 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr9_+_31222026 | 0.67 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr2_+_40294313 | 0.66 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr1_+_11107688 | 0.66 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr23_-_40194732 | 0.66 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr8_-_44223899 | 0.64 |
ENSDART00000143020
|
stx2b
|
syntaxin 2b |
| chr25_+_5015019 | 0.64 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr24_+_28383278 | 0.64 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr18_+_38775277 | 0.63 |
ENSDART00000186129
|
fam214a
|
family with sequence similarity 214, member A |
| chr14_+_6963312 | 0.62 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr17_+_8799661 | 0.62 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr22_+_18389271 | 0.61 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr14_+_29609245 | 0.59 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr13_-_48388726 | 0.59 |
ENSDART00000169473
|
fbxo11a
|
F-box protein 11a |
| chr8_-_44223473 | 0.59 |
ENSDART00000098525
|
stx2b
|
syntaxin 2b |
| chr4_+_13931733 | 0.58 |
ENSDART00000141742
ENSDART00000067175 |
pphln1
|
periphilin 1 |
| chr15_-_43238220 | 0.58 |
ENSDART00000027019
|
wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr8_-_8489685 | 0.58 |
ENSDART00000131849
ENSDART00000064113 |
abt1
|
activator of basal transcription 1 |
| chr15_+_19990068 | 0.57 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr1_-_26045560 | 0.57 |
ENSDART00000172737
ENSDART00000076120 ENSDART00000193593 |
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr5_+_32490238 | 0.56 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr22_-_3182965 | 0.56 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr7_-_17814118 | 0.56 |
ENSDART00000179688
|
ecsit
|
ECSIT signalling integrator |
| chr21_-_3700334 | 0.55 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr9_-_53537989 | 0.54 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr3_-_50139860 | 0.53 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr25_+_4635355 | 0.53 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr11_-_3897067 | 0.52 |
ENSDART00000134858
|
rpn1
|
ribophorin I |
| chr17_+_8799451 | 0.52 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr18_+_45542981 | 0.52 |
ENSDART00000140357
|
kifc3
|
kinesin family member C3 |
| chr22_+_22438783 | 0.52 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr16_+_48460873 | 0.51 |
ENSDART00000159902
|
ext1a
|
exostosin glycosyltransferase 1a |
| chr24_+_28561038 | 0.50 |
ENSDART00000147063
|
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr18_-_16315822 | 0.50 |
ENSDART00000136479
|
rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr14_+_31529958 | 0.49 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr8_-_8489886 | 0.49 |
ENSDART00000183334
|
abt1
|
activator of basal transcription 1 |
| chr18_+_36782930 | 0.48 |
ENSDART00000004129
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr23_-_33680265 | 0.48 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr4_-_5370468 | 0.48 |
ENSDART00000133423
ENSDART00000184963 |
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr23_+_38957738 | 0.47 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr14_-_7375049 | 0.47 |
ENSDART00000054809
|
trappc11
|
trafficking protein particle complex 11 |
| chr10_+_37500234 | 0.47 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr13_+_21919786 | 0.46 |
ENSDART00000182440
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr16_+_21918503 | 0.45 |
ENSDART00000167919
|
znf687a
|
zinc finger protein 687a |
| chr11_+_2902592 | 0.45 |
ENSDART00000093108
|
kif21b
|
kinesin family member 21B |
| chr13_-_4018888 | 0.45 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr12_-_4408828 | 0.45 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr3_-_36152974 | 0.45 |
ENSDART00000002568
|
engase
|
endo-beta-N-acetylglucosaminidase |
| chr14_+_7902374 | 0.44 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr18_-_14901437 | 0.44 |
ENSDART00000145842
ENSDART00000008035 |
trabd
|
TraB domain containing |
| chr17_-_27266053 | 0.44 |
ENSDART00000110903
|
E2F2
|
si:ch211-160f23.5 |
| chr7_-_30544702 | 0.44 |
ENSDART00000182409
|
sltm
|
SAFB-like, transcription modulator |
| chr8_+_25892319 | 0.43 |
ENSDART00000187167
ENSDART00000078163 |
tmem115
|
transmembrane protein 115 |
| chr7_+_38750871 | 0.43 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr22_+_11144153 | 0.42 |
ENSDART00000047442
|
bcor
|
BCL6 corepressor |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax10+pax6a+pax6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.7 | 2.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 2.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.4 | 1.7 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.4 | 1.7 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.4 | 9.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.4 | 2.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 3.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 1.1 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.3 | 1.9 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 1.6 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.3 | 1.3 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.3 | 1.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 2.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.7 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 2.5 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 1.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 2.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 1.6 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 3.3 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.3 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.2 | 0.6 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 2.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.5 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 2.8 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 3.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 1.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.7 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 1.1 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 1.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.9 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 1.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.5 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 2.5 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 4.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.7 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 2.2 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.1 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.7 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.6 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.5 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.4 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 3.2 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 2.8 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 2.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 1.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.9 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 1.1 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 0.6 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.8 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 1.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 16.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 2.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 2.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 0.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 2.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 2.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 1.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.7 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 1.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 2.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 4.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 3.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.6 | 9.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.5 | 16.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.5 | 2.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 1.9 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 2.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 2.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.3 | 3.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 2.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 3.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.7 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.2 | 1.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 1.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 2.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 2.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 3.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 0.6 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.6 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 2.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 3.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 2.3 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 1.3 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |