Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for osr1
Z-value: 1.05
Transcription factors associated with osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr1
|
ENSDARG00000014091 | odd-skipped related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| osr1 | dr11_v1_chr13_+_32148338_32148338 | 0.89 | 6.7e-07 | Click! |
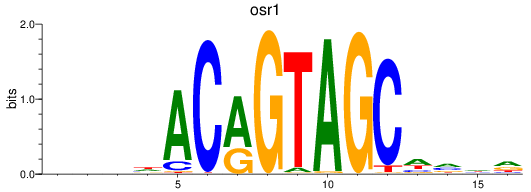
Activity profile of osr1 motif
Sorted Z-values of osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_34750169 | 4.19 |
ENSDART00000149380
|
si:dkey-151m22.8
|
si:dkey-151m22.8 |
| chr2_-_49557932 | 4.00 |
ENSDART00000141902
|
si:ch211-209f23.3
|
si:ch211-209f23.3 |
| chr5_-_61787969 | 3.50 |
ENSDART00000112744
|
gas2l2
|
growth arrest specific 2 like 2 |
| chr3_+_15805917 | 3.33 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr18_-_29961784 | 3.00 |
ENSDART00000135601
ENSDART00000140661 |
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr2_-_43942595 | 2.81 |
ENSDART00000147925
|
si:ch211-195h23.4
|
si:ch211-195h23.4 |
| chr2_+_49799470 | 2.77 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr7_+_24391129 | 2.77 |
ENSDART00000108753
|
poln
|
polymerase (DNA directed) nu |
| chr18_+_46382484 | 2.65 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr10_+_2587234 | 2.54 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr12_-_2993095 | 2.37 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr21_+_4702773 | 2.36 |
ENSDART00000147404
|
si:dkey-102g19.3
|
si:dkey-102g19.3 |
| chr22_-_28871787 | 2.28 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr5_-_36949476 | 2.20 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr23_-_42752550 | 2.17 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr23_-_42752387 | 2.15 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr4_-_8040436 | 2.14 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr14_+_4151379 | 1.95 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr2_+_56463167 | 1.92 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr22_-_21314821 | 1.91 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr20_-_2667902 | 1.90 |
ENSDART00000036373
|
cfap206
|
cilia and flagella associated protein 206 |
| chr8_-_24332284 | 1.86 |
ENSDART00000163241
|
pimr142
|
Pim proto-oncogene, serine/threonine kinase, related 142 |
| chr2_+_22531185 | 1.85 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr7_-_41295511 | 1.85 |
ENSDART00000173455
|
si:dkey-86l18.8
|
si:dkey-86l18.8 |
| chr7_-_52388090 | 1.71 |
ENSDART00000174104
|
wdr93
|
WD repeat domain 93 |
| chr8_-_24502997 | 1.69 |
ENSDART00000184801
ENSDART00000179794 |
BX296549.1
|
|
| chr20_-_29787192 | 1.69 |
ENSDART00000125348
ENSDART00000048759 |
id2b
|
inhibitor of DNA binding 2b |
| chr25_+_32530976 | 1.68 |
ENSDART00000156190
ENSDART00000103324 |
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr2_-_24898226 | 1.65 |
ENSDART00000134936
|
si:dkey-149i17.9
|
si:dkey-149i17.9 |
| chr11_+_26604224 | 1.65 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr7_+_26534131 | 1.63 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr19_+_31576849 | 1.60 |
ENSDART00000134107
ENSDART00000088401 |
acot13
|
acyl-CoA thioesterase 13 |
| chr7_-_52388734 | 1.59 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr15_-_32383529 | 1.56 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr2_-_37896965 | 1.55 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr8_-_36554675 | 1.55 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr16_-_7214513 | 1.52 |
ENSDART00000150005
|
si:ch73-341k19.1
|
si:ch73-341k19.1 |
| chr6_+_46431848 | 1.48 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr12_-_38548299 | 1.48 |
ENSDART00000153374
|
si:dkey-1f1.3
|
si:dkey-1f1.3 |
| chr8_-_24697586 | 1.47 |
ENSDART00000188639
|
BX324132.4
|
|
| chr14_+_17137023 | 1.46 |
ENSDART00000080712
|
slc43a3b
|
solute carrier family 43, member 3b |
| chr7_+_44713135 | 1.44 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr15_-_39848822 | 1.44 |
ENSDART00000155230
|
si:dkey-263j23.1
|
si:dkey-263j23.1 |
| chr10_-_25860102 | 1.44 |
ENSDART00000080789
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr8_-_24366726 | 1.43 |
ENSDART00000182351
ENSDART00000187099 ENSDART00000181534 |
CR450805.1
|
|
| chr24_-_9960290 | 1.42 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr13_+_9213805 | 1.42 |
ENSDART00000131455
|
pimr158
|
Pim proto-oncogene, serine/threonine kinase, related 158 |
| chr14_-_24391424 | 1.38 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr5_+_26847190 | 1.38 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr1_+_44767657 | 1.36 |
ENSDART00000073717
|
si:dkey-9i23.4
|
si:dkey-9i23.4 |
| chr5_+_69747417 | 1.35 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr20_+_1412193 | 1.35 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr3_-_30123113 | 1.34 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr2_+_3986083 | 1.33 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr8_+_50493095 | 1.32 |
ENSDART00000108655
ENSDART00000134382 |
si:dkey-263f15.2
|
si:dkey-263f15.2 |
| chr3_-_37758487 | 1.32 |
ENSDART00000150938
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr14_+_21038462 | 1.31 |
ENSDART00000162999
|
pimr193
|
Pim proto-oncogene, serine/threonine kinase, related 193 |
| chr6_-_52723901 | 1.31 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr10_-_34089779 | 1.28 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr3_-_16285861 | 1.25 |
ENSDART00000140103
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr3_-_29899172 | 1.24 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr4_+_3287819 | 1.23 |
ENSDART00000168633
|
CABZ01085700.1
|
|
| chr7_+_29954709 | 1.23 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr16_+_53489676 | 1.21 |
ENSDART00000074653
|
grinab
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1b (glutamate binding) |
| chr7_-_27033080 | 1.21 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr3_-_45298487 | 1.20 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr10_+_38512270 | 1.20 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr8_+_4798158 | 1.19 |
ENSDART00000031650
|
hsp70l
|
heat shock cognate 70-kd protein, like |
| chr6_+_49082796 | 1.15 |
ENSDART00000182446
|
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr14_+_35901249 | 1.08 |
ENSDART00000105604
|
zgc:77938
|
zgc:77938 |
| chr10_-_34107919 | 1.03 |
ENSDART00000167428
ENSDART00000145279 |
pimr144
pimr146
|
Pim proto-oncogene, serine/threonine kinase, related 144 Pim proto-oncogene, serine/threonine kinase, related 146 |
| chr10_-_34223567 | 1.03 |
ENSDART00000184889
|
pimr145
|
Pim proto-oncogene, serine/threonine kinase, related 145 |
| chr23_+_7379728 | 1.02 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr8_-_50981175 | 1.00 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr21_+_33454147 | 1.00 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr5_-_30978381 | 0.96 |
ENSDART00000127787
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr20_+_16881883 | 0.95 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr20_+_53521648 | 0.93 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr10_-_34067286 | 0.93 |
ENSDART00000140730
|
BX548044.8
|
|
| chr3_-_55139127 | 0.93 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr14_+_29769336 | 0.92 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr23_+_4324625 | 0.87 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr4_-_67969695 | 0.86 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr25_+_7670683 | 0.86 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr12_+_36413886 | 0.86 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr19_+_26714949 | 0.85 |
ENSDART00000188540
|
zgc:100906
|
zgc:100906 |
| chr23_+_24604481 | 0.85 |
ENSDART00000146332
|
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr2_-_20574193 | 0.85 |
ENSDART00000190448
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr21_-_25250594 | 0.85 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr2_-_24962820 | 0.83 |
ENSDART00000182767
|
hltf
|
helicase-like transcription factor |
| chr5_-_48307804 | 0.82 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr9_-_306515 | 0.82 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr18_+_45676788 | 0.81 |
ENSDART00000109948
ENSDART00000179864 |
qser1
|
glutamine and serine rich 1 |
| chr7_+_22767678 | 0.81 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr8_-_36469117 | 0.80 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr1_+_52563298 | 0.80 |
ENSDART00000142465
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr8_-_2506327 | 0.79 |
ENSDART00000101125
ENSDART00000125124 |
rpl6
|
ribosomal protein L6 |
| chr13_-_46917098 | 0.78 |
ENSDART00000183453
|
slc29a1a
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1a |
| chr9_-_12424231 | 0.77 |
ENSDART00000188952
|
zgc:162707
|
zgc:162707 |
| chr1_-_31657854 | 0.73 |
ENSDART00000085309
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr19_-_26823647 | 0.73 |
ENSDART00000002464
|
neu1
|
neuraminidase 1 |
| chr6_-_40884453 | 0.73 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr4_-_1824836 | 0.72 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr2_-_30668580 | 0.72 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr14_-_26536504 | 0.72 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr17_-_22021213 | 0.72 |
ENSDART00000078843
|
zmp:0000001102
|
zmp:0000001102 |
| chr7_+_17374560 | 0.71 |
ENSDART00000172272
ENSDART00000073467 |
nitr7a
|
novel immune-type receptor 7a |
| chr12_-_7824291 | 0.71 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr18_+_20869923 | 0.70 |
ENSDART00000138471
|
pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr4_+_47257854 | 0.70 |
ENSDART00000173868
|
crestin
|
crestin |
| chr15_-_576135 | 0.70 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr23_-_31403668 | 0.69 |
ENSDART00000147498
|
CR759830.1
|
|
| chr18_-_46258612 | 0.68 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr11_-_24347644 | 0.68 |
ENSDART00000089777
|
si:ch211-15p9.2
|
si:ch211-15p9.2 |
| chr4_-_6567355 | 0.67 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr16_-_28876479 | 0.67 |
ENSDART00000047862
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr8_-_7475917 | 0.66 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr5_-_57169604 | 0.66 |
ENSDART00000136232
|
si:dkey-79c1.1
|
si:dkey-79c1.1 |
| chr5_+_69808763 | 0.66 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr4_-_12323228 | 0.66 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr8_-_25728628 | 0.65 |
ENSDART00000127237
|
foxp3a
|
forkhead box P3a |
| chr13_-_32635859 | 0.64 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr5_+_13373593 | 0.64 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr8_+_48858132 | 0.63 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr19_+_10794072 | 0.63 |
ENSDART00000151518
ENSDART00000142614 |
lipea
|
lipase, hormone-sensitive a |
| chr14_-_40390757 | 0.63 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr11_+_14199802 | 0.62 |
ENSDART00000102520
ENSDART00000133172 |
palm1a
|
paralemmin 1a |
| chr6_+_52947186 | 0.61 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr3_-_16784280 | 0.61 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr7_+_3872189 | 0.61 |
ENSDART00000064221
|
si:dkey-88n24.5
|
si:dkey-88n24.5 |
| chr13_-_45523026 | 0.59 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr22_-_11626014 | 0.58 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr18_+_7204378 | 0.58 |
ENSDART00000142905
|
vwf
|
von Willebrand factor |
| chr12_-_18898413 | 0.58 |
ENSDART00000181281
ENSDART00000121866 |
desi1b
|
desumoylating isopeptidase 1b |
| chr18_+_30507839 | 0.57 |
ENSDART00000026866
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr15_-_32383340 | 0.57 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr22_-_16658996 | 0.57 |
ENSDART00000138429
|
si:dkey-38n4.2
|
si:dkey-38n4.2 |
| chr18_-_977075 | 0.56 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr11_-_33960318 | 0.55 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr17_-_50422584 | 0.55 |
ENSDART00000157095
|
si:ch211-235i11.5
|
si:ch211-235i11.5 |
| chr1_+_23783349 | 0.54 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr7_-_18508815 | 0.54 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr7_+_48805534 | 0.54 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr10_+_19554604 | 0.54 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr13_+_35746440 | 0.54 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr11_-_36350876 | 0.53 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr7_-_17600415 | 0.52 |
ENSDART00000080717
|
nitr6b
|
novel immune-type receptor 6b |
| chr2_-_9748039 | 0.51 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr2_+_37897079 | 0.51 |
ENSDART00000141784
|
tep1
|
telomerase-associated protein 1 |
| chr9_-_18568927 | 0.50 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr10_+_33744098 | 0.50 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr5_+_30682737 | 0.50 |
ENSDART00000172886
|
pdzd3b
|
PDZ domain containing 3b |
| chr1_-_21723329 | 0.49 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr7_-_59564011 | 0.49 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr18_-_49116382 | 0.48 |
ENSDART00000174157
|
BX663503.3
|
|
| chr7_-_55292116 | 0.48 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr14_+_33955807 | 0.48 |
ENSDART00000173316
|
si:ch73-22o18.1
|
si:ch73-22o18.1 |
| chr23_+_43809484 | 0.48 |
ENSDART00000149534
|
si:ch211-149b19.3
|
si:ch211-149b19.3 |
| chr12_-_15620090 | 0.47 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr11_-_29996344 | 0.47 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr6_-_8498908 | 0.46 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr16_+_35344031 | 0.46 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr19_-_9503473 | 0.45 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr22_-_26323893 | 0.45 |
ENSDART00000105099
|
capn1b
|
calpain 1, (mu/I) large subunit b |
| chr25_+_35212919 | 0.45 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr7_-_17635717 | 0.45 |
ENSDART00000080704
|
nitr6b
|
novel immune-type receptor 6b |
| chr17_-_14451405 | 0.45 |
ENSDART00000146786
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr19_-_6134802 | 0.45 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr25_+_20119466 | 0.45 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr10_-_31016806 | 0.43 |
ENSDART00000027288
|
panx3
|
pannexin 3 |
| chr12_-_45669050 | 0.43 |
ENSDART00000124455
|
fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chr7_-_60156409 | 0.43 |
ENSDART00000006802
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr18_-_29977431 | 0.43 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr13_+_28417297 | 0.43 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr11_-_41130239 | 0.42 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr2_-_11512819 | 0.42 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr13_-_1408775 | 0.42 |
ENSDART00000049684
|
bag2
|
BCL2 associated athanogene 2 |
| chr21_+_45627775 | 0.42 |
ENSDART00000185466
|
irf1b
|
interferon regulatory factor 1b |
| chr14_-_13048355 | 0.41 |
ENSDART00000166434
|
si:dkey-35h6.1
|
si:dkey-35h6.1 |
| chr10_-_22506044 | 0.41 |
ENSDART00000100497
|
zgc:172339
|
zgc:172339 |
| chr24_+_10413484 | 0.41 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr14_+_8504410 | 0.40 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr3_-_15144067 | 0.39 |
ENSDART00000127738
ENSDART00000060426 ENSDART00000180799 |
fam173a
|
family with sequence similarity 173, member A |
| chr1_+_55643198 | 0.39 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr15_-_35960250 | 0.39 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr16_-_27749172 | 0.39 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr1_-_9134045 | 0.39 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr12_-_48467733 | 0.38 |
ENSDART00000153126
ENSDART00000152895 ENSDART00000014190 |
sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr20_+_53522059 | 0.38 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr7_-_38806186 | 0.38 |
ENSDART00000173669
|
atg13
|
ATG13 autophagy related 13 homolog (S. cerevisiae) |
| chr7_+_50464500 | 0.37 |
ENSDART00000191356
|
serinc4
|
serine incorporator 4 |
| chr10_+_21559605 | 0.37 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr2_-_22363460 | 0.37 |
ENSDART00000158486
|
selenof
|
selenoprotein F |
| chr6_+_52947699 | 0.37 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr7_+_31871830 | 0.36 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr1_-_40911332 | 0.35 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr13_+_18663600 | 0.34 |
ENSDART00000141647
|
selenou1a
|
selenoprotein U 1a |
| chr8_+_30709685 | 0.34 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr4_-_75158035 | 0.34 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr7_+_20656942 | 0.33 |
ENSDART00000100898
|
tnfsf12
|
TNF superfamily member 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 1.9 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.3 | 0.9 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.3 | 1.0 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 0.7 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 3.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.8 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 0.5 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 1.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 2.7 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 0.7 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.2 | 1.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 1.3 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 0.8 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 1.2 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 3.3 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 1.3 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 1.9 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.5 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.4 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 1.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.7 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 1.7 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.3 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 1.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 3.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.4 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.0 | 0.2 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) leukocyte apoptotic process(GO:0071887) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.8 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.7 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 1.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 2.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.2 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 2.1 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 9.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.2 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.8 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.8 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.6 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0030513 | bone mineralization(GO:0030282) positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.3 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.0 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.0 | 0.7 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 2.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.2 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 0.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 1.3 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0001534 | radial spoke(GO:0001534) |
| 0.5 | 1.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 1.2 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.2 | 1.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 3.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.8 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 5.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.2 | 3.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.7 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 1.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.2 | 1.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 0.5 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.2 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.6 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.7 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.3 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.9 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.7 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 5.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 3.3 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 2.8 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 3.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |