Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
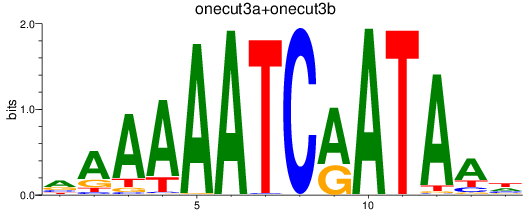
Results for onecut3a+onecut3b
Z-value: 0.47
Transcription factors associated with onecut3a+onecut3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut3a
|
ENSDARG00000056395 | one cut homeobox 3a |
|
onecut3b
|
ENSDARG00000091059 | one cut homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecut3a | dr11_v1_chr22_+_20461488_20461488 | 0.54 | 2.0e-02 | Click! |
| onecut3b | dr11_v1_chr2_+_57504073_57504073 | 0.26 | 2.9e-01 | Click! |
Activity profile of onecut3a+onecut3b motif
Sorted Z-values of onecut3a+onecut3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_39853788 | 1.25 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr5_-_3118346 | 1.17 |
ENSDART00000167554
|
hsf5
|
heat shock transcription factor family member 5 |
| chr5_-_57652168 | 1.11 |
ENSDART00000166245
|
gb:eh456644
|
expressed sequence EH456644 |
| chr15_+_21947117 | 1.09 |
ENSDART00000156234
|
ttc12
|
tetratricopeptide repeat domain 12 |
| chr6_-_40352215 | 1.07 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr21_-_40799404 | 1.06 |
ENSDART00000147405
ENSDART00000157512 |
zgc:162239
|
zgc:162239 |
| chr2_-_32505091 | 0.86 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr16_+_23960744 | 0.81 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr11_+_25259058 | 0.76 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr2_+_13462305 | 0.71 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr8_+_12925385 | 0.68 |
ENSDART00000085377
|
KIF2A
|
zgc:103670 |
| chr20_+_23440632 | 0.67 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr11_-_22605981 | 0.64 |
ENSDART00000186923
|
myog
|
myogenin |
| chr15_-_39948635 | 0.63 |
ENSDART00000114836
|
msh5
|
mutS homolog 5 |
| chr6_+_40714811 | 0.61 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr12_-_47793857 | 0.61 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr1_-_36151377 | 0.58 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr22_-_16494406 | 0.55 |
ENSDART00000062727
|
stx6
|
syntaxin 6 |
| chr16_+_32729223 | 0.54 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr6_+_46259950 | 0.54 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr2_-_42958619 | 0.53 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr18_+_33791608 | 0.51 |
ENSDART00000193991
|
si:dkey-145c18.2
|
si:dkey-145c18.2 |
| chr2_-_39759059 | 0.48 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr14_+_35748206 | 0.48 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr4_+_7827261 | 0.46 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr14_+_35748385 | 0.45 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr4_+_27018663 | 0.45 |
ENSDART00000180778
|
CR749777.1
|
|
| chr20_-_23171430 | 0.43 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr21_+_18877130 | 0.42 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr24_+_23730061 | 0.41 |
ENSDART00000080343
|
ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr17_+_23937262 | 0.40 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr18_-_6766354 | 0.40 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr11_+_25064519 | 0.40 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr16_-_12173554 | 0.38 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr11_-_10798021 | 0.37 |
ENSDART00000167112
ENSDART00000179725 ENSDART00000091923 ENSDART00000185825 |
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr21_-_41369370 | 0.37 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr22_-_7669624 | 0.37 |
ENSDART00000189617
|
AL929276.2
|
|
| chr5_-_67661102 | 0.37 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_-_28761454 | 0.35 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr19_-_18855513 | 0.35 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr23_+_16908012 | 0.34 |
ENSDART00000133946
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr16_-_54942532 | 0.33 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr3_-_28665291 | 0.33 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr15_+_19458982 | 0.33 |
ENSDART00000132665
|
zgc:77784
|
zgc:77784 |
| chr10_-_31782616 | 0.32 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr1_+_292545 | 0.31 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr16_-_12173399 | 0.30 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr16_+_19732543 | 0.30 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr12_-_13336703 | 0.30 |
ENSDART00000134356
|
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_+_19421841 | 0.29 |
ENSDART00000159090
ENSDART00000084771 |
pde9a
|
phosphodiesterase 9A |
| chr2_+_47581997 | 0.29 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr2_-_8611675 | 0.29 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr8_-_15292197 | 0.28 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr20_+_18740518 | 0.27 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr24_-_26995164 | 0.27 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr10_+_10801719 | 0.27 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr11_+_37201483 | 0.26 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr25_-_11088839 | 0.26 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr20_+_5074118 | 0.26 |
ENSDART00000104581
ENSDART00000142518 |
ccnk
|
cyclin K |
| chr11_+_24815667 | 0.26 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr1_+_40140498 | 0.25 |
ENSDART00000101640
|
zgc:152658
|
zgc:152658 |
| chr10_-_28835771 | 0.25 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr24_-_23320223 | 0.25 |
ENSDART00000135846
|
zfhx4
|
zinc finger homeobox 4 |
| chr11_+_6819050 | 0.25 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr19_-_18855717 | 0.24 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr15_-_34785594 | 0.24 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr4_-_21652812 | 0.23 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr23_-_28025943 | 0.23 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr3_+_32443395 | 0.23 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr21_-_43482426 | 0.22 |
ENSDART00000192901
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr7_+_40658003 | 0.22 |
ENSDART00000172435
|
lmbr1
|
limb development membrane protein 1 |
| chr13_+_24402406 | 0.22 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr11_+_24815927 | 0.21 |
ENSDART00000146838
|
rabif
|
RAB interacting factor |
| chr2_+_45159855 | 0.21 |
ENSDART00000056333
|
CU929150.1
|
|
| chr20_-_23440955 | 0.20 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr25_+_31267268 | 0.19 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr11_+_5817202 | 0.19 |
ENSDART00000126084
|
ctdspl3
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3 |
| chr16_-_47426482 | 0.19 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr9_+_32930622 | 0.19 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr16_-_44127307 | 0.18 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr2_-_59145027 | 0.18 |
ENSDART00000128320
|
FO834803.1
|
|
| chr20_+_18740799 | 0.18 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr2_-_45510699 | 0.18 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr6_+_3828560 | 0.18 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr16_+_33655890 | 0.18 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr10_-_7785930 | 0.17 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr19_+_25649626 | 0.16 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr21_-_39058490 | 0.16 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr17_-_45378473 | 0.16 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr7_+_63325819 | 0.16 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr4_-_18304932 | 0.16 |
ENSDART00000004222
|
plxnc1
|
plexin C1 |
| chr20_+_42978499 | 0.15 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr7_-_41693004 | 0.14 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr1_-_23157583 | 0.13 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr2_+_5887626 | 0.13 |
ENSDART00000147831
|
si:ch211-168b3.1
|
si:ch211-168b3.1 |
| chr2_-_56387041 | 0.13 |
ENSDART00000036240
|
cers4b
|
ceramide synthase 4b |
| chr17_-_25382367 | 0.13 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr20_+_52546186 | 0.13 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr22_+_19528851 | 0.13 |
ENSDART00000145079
|
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr1_+_23784905 | 0.13 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr12_+_48204891 | 0.13 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr17_-_14451405 | 0.12 |
ENSDART00000146786
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr8_+_9866351 | 0.12 |
ENSDART00000133985
|
kcnd1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr15_+_17441734 | 0.12 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr19_-_38539670 | 0.12 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr18_-_44847855 | 0.12 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr1_+_51391082 | 0.12 |
ENSDART00000063936
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr8_-_23081511 | 0.12 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr4_+_5842433 | 0.11 |
ENSDART00000124085
ENSDART00000179848 |
usp18
|
ubiquitin specific peptidase 18 |
| chr5_+_19309877 | 0.11 |
ENSDART00000190338
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr7_+_48675347 | 0.11 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr2_-_45510223 | 0.10 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr22_-_3564563 | 0.10 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr5_-_69212184 | 0.10 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr16_-_31525290 | 0.10 |
ENSDART00000109996
|
wisp1b
|
WNT1 inducible signaling pathway protein 1b |
| chr3_+_1492174 | 0.09 |
ENSDART00000112979
|
sox10
|
SRY (sex determining region Y)-box 10 |
| chr21_+_6212844 | 0.09 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr23_-_15216654 | 0.09 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr12_+_48841182 | 0.09 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr1_+_54069450 | 0.09 |
ENSDART00000108601
ENSDART00000187878 |
dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr18_+_15876385 | 0.09 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr24_-_39826865 | 0.09 |
ENSDART00000089232
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr14_+_49376011 | 0.08 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr16_-_2650341 | 0.08 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr17_-_8169774 | 0.08 |
ENSDART00000091828
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr9_+_51147380 | 0.08 |
ENSDART00000003913
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr19_+_11984725 | 0.08 |
ENSDART00000185960
|
spag1a
|
sperm associated antigen 1a |
| chr8_-_14484599 | 0.08 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr15_+_11381532 | 0.08 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr17_-_5424175 | 0.07 |
ENSDART00000171063
ENSDART00000188102 |
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr15_+_17446796 | 0.07 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr16_-_26435431 | 0.07 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr2_+_47582488 | 0.06 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr20_-_13660767 | 0.06 |
ENSDART00000127654
|
TAGAP
|
si:ch211-122h15.4 |
| chr13_-_37383625 | 0.06 |
ENSDART00000192888
ENSDART00000100322 ENSDART00000138934 |
kcnh5b
|
potassium voltage-gated channel, subfamily H (eag-related), member 5b |
| chr3_+_39958407 | 0.06 |
ENSDART00000099754
|
ccdc97
|
coiled-coil domain containing 97 |
| chr20_-_3004126 | 0.06 |
ENSDART00000064390
|
ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr2_+_22042745 | 0.05 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr4_-_76488854 | 0.05 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr5_+_3118231 | 0.05 |
ENSDART00000179809
|
supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr20_+_16639848 | 0.05 |
ENSDART00000063944
ENSDART00000152359 |
tmem30ab
|
transmembrane protein 30Ab |
| chr4_+_8016457 | 0.05 |
ENSDART00000014036
|
optn
|
optineurin |
| chr23_-_35195908 | 0.05 |
ENSDART00000122429
|
klf15
|
Kruppel-like factor 15 |
| chr5_+_56026031 | 0.05 |
ENSDART00000050970
|
fzd9a
|
frizzled class receptor 9a |
| chr25_-_13703826 | 0.04 |
ENSDART00000163398
|
pla2g15
|
phospholipase A2, group XV |
| chr12_-_35582683 | 0.04 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr15_-_34418525 | 0.04 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr16_+_22587661 | 0.04 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr3_+_32689707 | 0.03 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr20_+_22130284 | 0.03 |
ENSDART00000025575
|
clocka
|
clock circadian regulator a |
| chr5_+_42136359 | 0.03 |
ENSDART00000083731
|
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr7_-_4461104 | 0.03 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr17_-_45247332 | 0.03 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr1_+_19332837 | 0.03 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr23_+_28770225 | 0.03 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr18_-_20560007 | 0.02 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr2_+_6243144 | 0.02 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr10_-_44306636 | 0.02 |
ENSDART00000191068
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr20_-_13660600 | 0.02 |
ENSDART00000063826
|
TAGAP
|
si:ch211-122h15.4 |
| chr6_-_33707278 | 0.02 |
ENSDART00000188103
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr21_-_28901095 | 0.02 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr12_-_43428542 | 0.02 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr2_-_17044959 | 0.01 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr16_+_3067134 | 0.01 |
ENSDART00000012048
|
cyb5r4
|
cytochrome b5 reductase 4 |
| chr12_-_18883079 | 0.01 |
ENSDART00000178619
|
shisa8b
|
shisa family member 8b |
| chr18_+_20560442 | 0.01 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr21_-_23475361 | 0.01 |
ENSDART00000156658
ENSDART00000157454 |
ncam1a
|
neural cell adhesion molecule 1a |
| chr3_+_23691847 | 0.01 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr7_-_24022340 | 0.01 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr7_-_36096582 | 0.01 |
ENSDART00000188507
|
CR848715.1
|
|
| chr1_-_21287724 | 0.00 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr15_-_34892664 | 0.00 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr22_-_8910567 | 0.00 |
ENSDART00000187896
ENSDART00000188511 ENSDART00000175764 |
FO393424.2
|
|
| chr19_-_46018152 | 0.00 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut3a+onecut3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 0.5 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.6 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.5 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.6 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 1.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.4 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 1.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.1 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.1 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.3 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 0.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.2 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |