Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
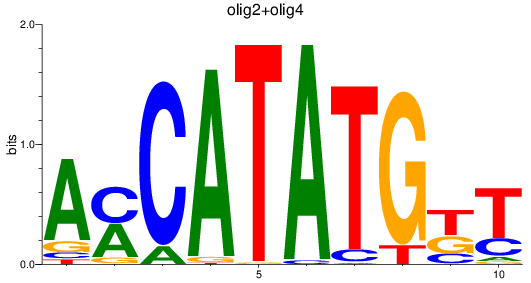
Results for olig3_olig2+olig4_olig1
Z-value: 1.85
Transcription factors associated with olig3_olig2+olig4_olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
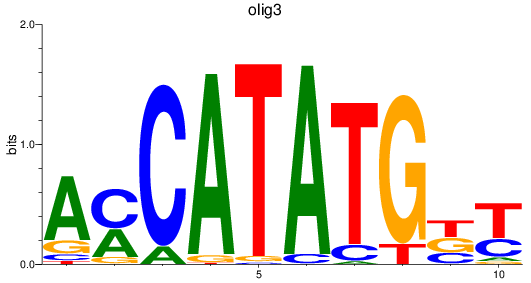
olig3
|
ENSDARG00000074253 | oligodendrocyte transcription factor 3 |
|
olig2
|
ENSDARG00000040946 | oligodendrocyte lineage transcription factor 2 |
|
olig4
|
ENSDARG00000052610 | oligodendrocyte transcription factor 4 |
|
olig4
|
ENSDARG00000116678 | oligodendrocyte transcription factor 4 |
|
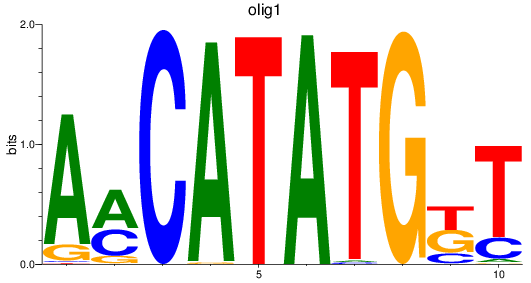
olig1
|
ENSDARG00000040948 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| olig4 | dr11_v1_chr13_+_45967179_45967179 | -0.92 | 4.7e-08 | Click! |
| olig1 | dr11_v1_chr9_-_32730487_32730487 | -0.52 | 2.7e-02 | Click! |
| olig2 | dr11_v1_chr9_-_32753535_32753535 | -0.31 | 2.2e-01 | Click! |
Activity profile of olig3_olig2+olig4_olig1 motif
Sorted Z-values of olig3_olig2+olig4_olig1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_46020508 | 6.83 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr1_-_43712120 | 6.62 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr16_-_39267185 | 6.22 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr12_-_17863467 | 6.16 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr17_-_15189397 | 5.57 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr19_-_8798178 | 5.49 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr18_-_40708537 | 5.25 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr8_+_1009831 | 5.03 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr19_-_12212692 | 4.49 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr3_+_43374571 | 4.33 |
ENSDART00000182497
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr20_+_52529044 | 4.30 |
ENSDART00000158230
ENSDART00000002787 |
pycr3
|
pyrroline-5-carboxylate reductase 3 |
| chr20_-_4031475 | 4.25 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr6_-_7749396 | 4.08 |
ENSDART00000105232
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr9_-_6502491 | 4.07 |
ENSDART00000102672
|
nck2a
|
NCK adaptor protein 2a |
| chr19_-_23249822 | 4.02 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr3_+_43373867 | 3.88 |
ENSDART00000159455
ENSDART00000172425 |
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr20_+_33924235 | 3.86 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr2_-_39036604 | 3.71 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr20_-_31252809 | 3.68 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr13_+_11550454 | 3.62 |
ENSDART00000034935
ENSDART00000166908 |
desi2
|
desumoylating isopeptidase 2 |
| chr17_-_17756344 | 3.42 |
ENSDART00000190146
|
adck1
|
aarF domain containing kinase 1 |
| chr19_+_38167468 | 3.41 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr3_+_7763114 | 3.37 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr7_+_9290929 | 3.33 |
ENSDART00000128530
|
snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr17_-_15188440 | 3.29 |
ENSDART00000151885
|
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr7_+_46019780 | 3.26 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr5_+_28259655 | 3.25 |
ENSDART00000087684
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr20_-_23876291 | 3.20 |
ENSDART00000043316
|
katna1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr3_+_43774369 | 3.19 |
ENSDART00000157964
|
zc3h7a
|
zinc finger CCCH-type containing 7A |
| chr3_+_19665319 | 3.19 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr19_-_27578929 | 3.18 |
ENSDART00000177368
|
si:dkeyp-46h3.3
|
si:dkeyp-46h3.3 |
| chr16_-_8132742 | 3.18 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr24_+_7741521 | 3.16 |
ENSDART00000009687
|
zgc:56095
|
zgc:56095 |
| chr2_+_1988036 | 3.12 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr14_+_35405518 | 3.11 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr5_-_25576462 | 2.97 |
ENSDART00000165147
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr5_-_29512538 | 2.96 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr5_-_22602979 | 2.95 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr2_-_42558549 | 2.91 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr1_+_40613297 | 2.88 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr9_+_8364553 | 2.87 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr20_-_34028967 | 2.86 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr24_-_16980337 | 2.81 |
ENSDART00000183812
|
klhl15
|
kelch-like family member 15 |
| chr10_-_26225548 | 2.80 |
ENSDART00000132019
ENSDART00000079194 |
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr5_+_37379825 | 2.80 |
ENSDART00000171826
|
klhl13
|
kelch-like family member 13 |
| chr22_-_11054244 | 2.78 |
ENSDART00000105823
|
insrb
|
insulin receptor b |
| chr2_-_10943093 | 2.77 |
ENSDART00000148999
|
ssbp3a
|
single stranded DNA binding protein 3a |
| chr19_+_7173613 | 2.74 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr24_-_41797681 | 2.71 |
ENSDART00000169643
|
arhgap28
|
Rho GTPase activating protein 28 |
| chr19_+_24324967 | 2.66 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr19_+_9113932 | 2.63 |
ENSDART00000060442
|
setdb1a
|
SET domain, bifurcated 1a |
| chr9_+_28150275 | 2.60 |
ENSDART00000192129
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr1_+_16600690 | 2.57 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr8_+_47219107 | 2.56 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr13_+_22731356 | 2.56 |
ENSDART00000133064
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr5_-_32505276 | 2.55 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr14_-_34633960 | 2.52 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr18_-_3520358 | 2.50 |
ENSDART00000181412
|
capn5a
|
calpain 5a |
| chr13_+_11439486 | 2.47 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr13_+_21676235 | 2.46 |
ENSDART00000137804
ENSDART00000134950 ENSDART00000129653 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr20_-_14781904 | 2.37 |
ENSDART00000187200
ENSDART00000179912 ENSDART00000160481 ENSDART00000026969 |
suco
|
SUN domain containing ossification factor |
| chr20_+_28803642 | 2.35 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr15_+_4988189 | 2.35 |
ENSDART00000142995
ENSDART00000062852 |
spcs2
|
signal peptidase complex subunit 2 |
| chr11_+_25693395 | 2.34 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr25_-_37338048 | 2.33 |
ENSDART00000073439
|
trim44
|
tripartite motif containing 44 |
| chr21_+_13387965 | 2.33 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr3_-_27647845 | 2.32 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr10_-_4961923 | 2.31 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr23_-_4019699 | 2.29 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr9_-_11676491 | 2.27 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr23_-_18057270 | 2.26 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr14_+_31618982 | 2.26 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr1_-_22652424 | 2.23 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr7_+_38667066 | 2.23 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr19_-_7406933 | 2.22 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr20_+_34390196 | 2.21 |
ENSDART00000183596
|
trmt1l
|
tRNA methyltransferase 1-like |
| chr8_-_51507144 | 2.19 |
ENSDART00000024882
ENSDART00000135166 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr1_+_49415281 | 2.17 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_+_15581270 | 2.16 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr9_-_25328527 | 2.13 |
ENSDART00000060840
|
med4
|
mediator complex subunit 4 |
| chr13_+_30421472 | 2.11 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr13_+_22698215 | 2.09 |
ENSDART00000137467
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr22_-_14272699 | 2.07 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr16_-_17200120 | 2.07 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr24_-_10828560 | 2.03 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr19_+_10661520 | 2.01 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr8_+_36582728 | 2.00 |
ENSDART00000049230
|
pqbp1
|
polyglutamine binding protein 1 |
| chr19_-_46037835 | 1.99 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr1_+_19649545 | 1.97 |
ENSDART00000054575
|
tmem192
|
transmembrane protein 192 |
| chr11_-_279328 | 1.96 |
ENSDART00000066179
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr13_+_9368621 | 1.96 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr12_-_9498060 | 1.94 |
ENSDART00000160622
|
si:ch211-207i20.2
|
si:ch211-207i20.2 |
| chr19_+_32456974 | 1.93 |
ENSDART00000088265
|
atxn1a
|
ataxin 1a |
| chr2_-_45154550 | 1.91 |
ENSDART00000148595
ENSDART00000150002 |
capn10
|
calpain 10 |
| chr10_-_32465462 | 1.91 |
ENSDART00000134056
|
uvrag
|
UV radiation resistance associated gene |
| chr16_-_17586883 | 1.89 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr18_-_40913294 | 1.89 |
ENSDART00000059196
ENSDART00000098878 |
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr22_-_8006342 | 1.89 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr15_-_28587490 | 1.87 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr11_+_5588122 | 1.87 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr12_-_22355430 | 1.86 |
ENSDART00000153296
ENSDART00000056919 ENSDART00000159036 |
nsfb
|
N-ethylmaleimide-sensitive factor b |
| chr4_+_9178913 | 1.85 |
ENSDART00000168558
|
nfyba
|
nuclear transcription factor Y, beta a |
| chr24_-_16979728 | 1.85 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr20_-_48898560 | 1.84 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr20_+_28803977 | 1.79 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr9_+_48123002 | 1.78 |
ENSDART00000099794
ENSDART00000169733 |
klhl23
|
kelch-like family member 23 |
| chr6_-_42949184 | 1.76 |
ENSDART00000147208
|
edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr25_+_3294150 | 1.76 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr13_-_51922290 | 1.75 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr20_-_1265562 | 1.74 |
ENSDART00000189866
|
lats1
|
large tumor suppressor kinase 1 |
| chr6_+_21992820 | 1.73 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr25_+_19870603 | 1.73 |
ENSDART00000047251
|
gramd4b
|
GRAM domain containing 4b |
| chr5_-_39805874 | 1.72 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr23_-_31648026 | 1.71 |
ENSDART00000133569
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_28480546 | 1.71 |
ENSDART00000057696
ENSDART00000160858 |
git1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr14_+_10981703 | 1.70 |
ENSDART00000091169
|
abcb7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr2_-_57110477 | 1.70 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr17_-_8692722 | 1.69 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr21_-_2042037 | 1.68 |
ENSDART00000171131
ENSDART00000160144 |
add1
|
adducin 1 (alpha) |
| chr1_-_6028876 | 1.68 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr20_-_9123296 | 1.66 |
ENSDART00000188495
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr17_-_45104750 | 1.66 |
ENSDART00000075520
|
aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr9_-_30264415 | 1.66 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr23_+_17522867 | 1.66 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr16_+_53252951 | 1.64 |
ENSDART00000126543
|
si:ch211-269k10.5
|
si:ch211-269k10.5 |
| chr3_+_25907266 | 1.64 |
ENSDART00000170324
ENSDART00000192633 |
tom1
|
target of myb1 membrane trafficking protein |
| chr2_+_24770435 | 1.63 |
ENSDART00000078854
|
mpv17l2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr2_+_44972720 | 1.63 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr3_-_26184018 | 1.61 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr17_-_43594864 | 1.60 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr11_-_24532988 | 1.60 |
ENSDART00000067078
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr15_-_43270889 | 1.60 |
ENSDART00000166805
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr6_+_50393047 | 1.58 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr19_+_42227400 | 1.57 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr5_-_57289872 | 1.55 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr18_+_6866276 | 1.55 |
ENSDART00000187516
|
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr25_+_3104959 | 1.55 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr5_+_50869091 | 1.55 |
ENSDART00000083294
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr13_+_29926631 | 1.53 |
ENSDART00000135265
|
cuedc2
|
CUE domain containing 2 |
| chr22_-_37797695 | 1.52 |
ENSDART00000085931
ENSDART00000185443 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr23_+_7548797 | 1.52 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr17_-_24820801 | 1.51 |
ENSDART00000156606
|
trmt61b
|
tRNA methyltransferase 61B |
| chr5_+_44805269 | 1.51 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr24_-_7777389 | 1.50 |
ENSDART00000138541
|
rpgrip1
|
RPGR interacting protein 1 |
| chr3_-_54500354 | 1.49 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr12_-_3077395 | 1.49 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr22_-_37796998 | 1.48 |
ENSDART00000124742
ENSDART00000191232 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr9_-_32300783 | 1.48 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr10_+_15025006 | 1.48 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr5_+_44805028 | 1.47 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr4_-_4592287 | 1.47 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr5_+_47882319 | 1.47 |
ENSDART00000149316
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr1_-_55263736 | 1.46 |
ENSDART00000152504
ENSDART00000152687 |
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr1_+_14454663 | 1.46 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr5_+_52625975 | 1.46 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr6_+_23026170 | 1.44 |
ENSDART00000186683
|
srp68
|
signal recognition particle 68 |
| chr7_-_24520866 | 1.44 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr3_+_52953489 | 1.44 |
ENSDART00000125136
|
dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr20_+_25486021 | 1.44 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr8_-_53991184 | 1.44 |
ENSDART00000180675
|
SLC35E2
|
solute carrier family 35 member E2 |
| chr14_-_33425170 | 1.44 |
ENSDART00000124629
ENSDART00000105800 ENSDART00000001318 |
nkap
|
NFKB activating protein |
| chr7_+_72630369 | 1.43 |
ENSDART00000170698
|
FO905040.1
|
|
| chr1_+_39995008 | 1.43 |
ENSDART00000166251
|
aip
|
aryl hydrocarbon receptor interacting protein |
| chr17_+_15534815 | 1.42 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr1_-_34450784 | 1.42 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr17_+_28005763 | 1.42 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr5_+_20255568 | 1.42 |
ENSDART00000153643
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr21_+_20386865 | 1.42 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr20_-_3997531 | 1.41 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr21_+_974555 | 1.41 |
ENSDART00000130648
|
scarb2c
|
scavenger receptor class B, member 2c |
| chr24_-_9689915 | 1.40 |
ENSDART00000185972
ENSDART00000093046 |
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr13_-_29980215 | 1.40 |
ENSDART00000042049
|
hif1an
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr1_-_34450622 | 1.37 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr9_-_32300611 | 1.37 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr20_-_43743700 | 1.36 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr24_-_26885897 | 1.36 |
ENSDART00000180512
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr20_+_54333774 | 1.36 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr25_+_37397031 | 1.35 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr8_-_410728 | 1.35 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr8_-_26792912 | 1.35 |
ENSDART00000139787
|
kazna
|
kazrin, periplakin interacting protein a |
| chr12_-_20616160 | 1.35 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr19_+_42071033 | 1.34 |
ENSDART00000169067
ENSDART00000183436 |
nfyc
|
nuclear transcription factor Y, gamma |
| chr12_+_5048044 | 1.32 |
ENSDART00000161548
ENSDART00000172607 |
kif22
|
kinesin family member 22 |
| chr5_+_44804791 | 1.32 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr24_-_21172122 | 1.32 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr24_-_9989634 | 1.32 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr9_-_43644261 | 1.31 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr14_-_47314011 | 1.31 |
ENSDART00000178523
|
fstl5
|
follistatin-like 5 |
| chr2_-_4787566 | 1.30 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr10_-_2942900 | 1.30 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr10_+_18877362 | 1.29 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr5_+_32817688 | 1.29 |
ENSDART00000139472
|
crata
|
carnitine O-acetyltransferase a |
| chr21_-_1635268 | 1.29 |
ENSDART00000151258
|
zgc:152948
|
zgc:152948 |
| chr14_+_45028062 | 1.29 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr7_-_50410524 | 1.28 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr21_+_21195487 | 1.28 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr10_-_24759616 | 1.28 |
ENSDART00000079528
|
ilk
|
integrin-linked kinase |
| chr5_+_30518036 | 1.28 |
ENSDART00000161836
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr1_-_18585046 | 1.27 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr20_-_9123052 | 1.27 |
ENSDART00000125133
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of olig3_olig2+olig4_olig1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.0 | 4.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.9 | 7.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.9 | 2.6 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.8 | 4.1 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.7 | 2.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.7 | 2.2 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.7 | 2.8 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.7 | 2.7 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.7 | 2.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.6 | 1.9 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.6 | 1.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.5 | 1.6 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.5 | 1.5 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.5 | 4.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.5 | 3.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.4 | 1.7 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.4 | 2.9 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 1.2 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.4 | 1.6 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.4 | 0.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.4 | 3.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.4 | 2.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 2.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.4 | 1.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.4 | 1.5 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.4 | 1.9 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.4 | 1.5 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 1.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.3 | 1.0 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.3 | 2.0 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.3 | 1.0 | GO:0070228 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.3 | 1.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.3 | 1.3 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.3 | 0.9 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 3.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 4.0 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.3 | 5.5 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.3 | 1.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 1.4 | GO:0097201 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.3 | 1.7 | GO:0006574 | thymine catabolic process(GO:0006210) valine catabolic process(GO:0006574) thymine metabolic process(GO:0019859) |
| 0.3 | 0.8 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.3 | 1.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 4.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.3 | 1.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 3.4 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 1.3 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.3 | 3.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 3.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 2.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 0.7 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 1.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 3.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 1.7 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 1.2 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 0.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 2.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.2 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 0.8 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 0.6 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.2 | 0.6 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 2.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 1.4 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.2 | 2.0 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 1.2 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 0.8 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.2 | 9.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 1.9 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 0.7 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.3 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.2 | 0.7 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 2.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 3.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 2.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 0.7 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 1.8 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 2.0 | GO:0003139 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.2 | 1.0 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.2 | 0.8 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.2 | 0.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 1.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 1.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.5 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.7 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.9 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 1.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 3.2 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.1 | 2.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 2.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.8 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 2.7 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 0.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.4 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 3.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.6 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 1.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 2.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 2.6 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 2.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.3 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 1.2 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 3.9 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 0.7 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 2.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 1.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.3 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.2 | GO:0045830 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.9 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.4 | GO:0031649 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 2.7 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.1 | 0.7 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 3.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.1 | 1.1 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 0.4 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.1 | 0.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 1.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 2.0 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.3 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 7.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 3.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 4.4 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.1 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 4.2 | GO:0051170 | nuclear import(GO:0051170) |
| 0.1 | 3.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.1 | 0.5 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 2.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.5 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.5 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.7 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.2 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 4.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 4.0 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.0 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.5 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.8 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 2.2 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 1.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.1 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.0 | 0.3 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.5 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.3 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 1.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.3 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 1.3 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.6 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.3 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.7 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.6 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 2.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 1.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 1.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 9.1 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.7 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 2.2 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 2.4 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.5 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0030817 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 1.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.9 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.7 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.8 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 2.1 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 1.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 1.0 | 4.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.8 | 4.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 3.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.6 | 2.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 1.5 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.5 | 1.4 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.4 | 3.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 3.0 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.4 | 1.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.4 | 2.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 1.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.3 | 2.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 2.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 3.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 0.8 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.3 | 3.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.3 | 8.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.3 | 2.0 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.4 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 1.7 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 6.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 1.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 2.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 3.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 0.8 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 1.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.5 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 3.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 1.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 1.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 3.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 3.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0071256 | translocon complex(GO:0071256) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 7.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 15.5 | GO:0000323 | lytic vacuole(GO:0000323) |
| 0.1 | 0.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 3.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 2.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 13.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 1.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0098844 | dendritic spine head(GO:0044327) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 7.1 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 3.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.2 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.2 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.0 | 4.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.9 | 4.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.9 | 2.6 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.8 | 3.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.7 | 3.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.6 | 3.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.6 | 2.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.6 | 3.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.6 | 1.7 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.6 | 2.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.5 | 5.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.5 | 2.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.5 | 2.0 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.5 | 1.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.5 | 1.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.5 | 1.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.4 | 1.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.4 | 1.7 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.4 | 1.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.4 | 1.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.4 | 4.1 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 3.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 1.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 1.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 2.0 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.3 | 3.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 0.9 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 1.5 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.3 | 3.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 0.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 3.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 3.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 0.8 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 1.7 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.2 | 3.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 1.7 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 2.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.9 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 2.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 2.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 2.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 2.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 3.1 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.2 | 1.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 1.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 1.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.7 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 3.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 3.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 9.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 4.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 2.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.8 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.7 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 2.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.4 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.9 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 2.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 3.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.5 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 2.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.7 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.8 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.0 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.5 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.9 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.5 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 4.0 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 2.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.7 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 2.2 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 4.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 3.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 3.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 3.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 4.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 2.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 6.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.3 | GO:0042626 | P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 7.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 3.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 3.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 2.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 3.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 6.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 3.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 2.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 4.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.9 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |