Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
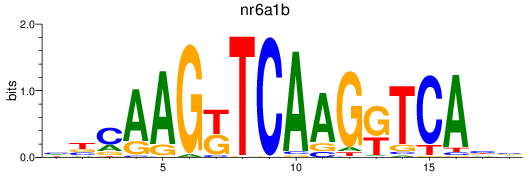
Results for nr6a1b
Z-value: 0.18
Transcription factors associated with nr6a1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1b
|
ENSDARG00000014480 | nuclear receptor subfamily 6, group A, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1b | dr11_v1_chr21_+_8198652_8198652 | 0.48 | 4.3e-02 | Click! |
Activity profile of nr6a1b motif
Sorted Z-values of nr6a1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10175898 | 0.45 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr9_-_7287375 | 0.18 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr12_+_33395748 | 0.18 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr8_-_54216499 | 0.18 |
ENSDART00000193678
|
mbd4
|
methyl-CpG binding domain protein 4 |
| chr2_+_24700922 | 0.17 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr25_+_7492663 | 0.16 |
ENSDART00000166496
|
cat
|
catalase |
| chr5_-_50992690 | 0.15 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr17_-_53348486 | 0.15 |
ENSDART00000161183
|
CABZ01066926.1
|
|
| chr22_+_24603930 | 0.14 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr11_+_45219558 | 0.14 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr8_-_49908978 | 0.14 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr3_-_40976288 | 0.13 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr3_+_12652244 | 0.12 |
ENSDART00000165980
|
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr3_-_40976463 | 0.11 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr6_+_39836474 | 0.10 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr25_+_17860798 | 0.10 |
ENSDART00000146845
|
pth1a
|
parathyroid hormone 1a |
| chr2_-_50225411 | 0.10 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr24_+_19542323 | 0.09 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr23_+_27779452 | 0.09 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr20_+_9128829 | 0.09 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr1_-_55118745 | 0.09 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr25_+_17860962 | 0.08 |
ENSDART00000163153
|
pth1a
|
parathyroid hormone 1a |
| chr1_-_11104805 | 0.08 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr5_+_6892195 | 0.08 |
ENSDART00000048201
|
gb:cr929477
|
expressed sequence CR929477 |
| chr7_-_31618166 | 0.08 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr1_-_49947290 | 0.07 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr9_+_24095677 | 0.07 |
ENSDART00000150443
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr13_+_713941 | 0.07 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr23_+_17102960 | 0.07 |
ENSDART00000053414
|
commd7
|
COMM domain containing 7 |
| chr12_+_30788912 | 0.06 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_-_47246948 | 0.06 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr16_+_48616220 | 0.06 |
ENSDART00000004751
ENSDART00000130045 |
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr1_+_57348756 | 0.05 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr10_-_16868211 | 0.05 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr18_+_8912113 | 0.05 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr3_+_22442445 | 0.05 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr6_-_30932078 | 0.04 |
ENSDART00000028612
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr14_-_25095808 | 0.04 |
ENSDART00000184244
|
matr3l1.1
|
matrin 3-like 1.1 |
| chr17_-_40956035 | 0.04 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr21_-_32374656 | 0.04 |
ENSDART00000112550
|
mapk9
|
mitogen-activated protein kinase 9 |
| chr6_+_48618512 | 0.04 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr15_+_22722684 | 0.04 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr6_+_41099787 | 0.04 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr3_+_19299309 | 0.04 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr7_-_15370042 | 0.03 |
ENSDART00000128183
|
ANPEP
|
si:ch211-276a23.5 |
| chr1_+_17527342 | 0.03 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr5_-_11971946 | 0.03 |
ENSDART00000166285
|
si:ch73-47f2.1
|
si:ch73-47f2.1 |
| chr20_+_4392687 | 0.03 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr1_-_49947482 | 0.03 |
ENSDART00000143398
|
sgms2
|
sphingomyelin synthase 2 |
| chr10_+_29260096 | 0.03 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr21_+_43669943 | 0.02 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr2_+_13694450 | 0.02 |
ENSDART00000077259
ENSDART00000189485 |
ebna1bp2
|
EBNA1 binding protein 2 |
| chr8_-_14091886 | 0.02 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr1_+_17527931 | 0.02 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr22_-_17781213 | 0.02 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr25_-_21609645 | 0.02 |
ENSDART00000087817
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr3_+_2669813 | 0.02 |
ENSDART00000014205
|
CR388047.1
|
|
| chr9_+_27371240 | 0.02 |
ENSDART00000060337
|
tlr20.3
|
toll-like receptor 20, tandem duplicate 3 |
| chr8_+_22472584 | 0.02 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr17_-_37054959 | 0.01 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr10_-_373575 | 0.01 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr9_+_32301456 | 0.01 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr7_+_29461060 | 0.01 |
ENSDART00000075721
|
foxb1b
|
forkhead box B1b |
| chr5_+_50879545 | 0.01 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr5_-_30475011 | 0.01 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr19_+_7878122 | 0.01 |
ENSDART00000131917
|
si:dkeyp-85e10.2
|
si:dkeyp-85e10.2 |
| chr23_-_19953089 | 0.01 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr16_-_45235947 | 0.01 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr4_-_34480599 | 0.01 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr11_-_12739532 | 0.01 |
ENSDART00000183734
|
AP1S2
|
adaptor related protein complex 1 sigma 2 subunit |
| chr4_-_41196398 | 0.01 |
ENSDART00000163401
|
si:dkeyp-82h4.3
|
si:dkeyp-82h4.3 |
| chr16_-_46558708 | 0.01 |
ENSDART00000128037
|
si:dkey-152b24.8
|
si:dkey-152b24.8 |
| chr23_-_19230627 | 0.01 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr13_-_22843562 | 0.01 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr7_-_30639385 | 0.01 |
ENSDART00000173618
|
myo1ea
|
myosin IE, a |
| chr3_-_49015203 | 0.01 |
ENSDART00000186138
|
BX769173.2
|
|
| chr25_+_33939728 | 0.01 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr1_+_29862074 | 0.01 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr15_+_19681718 | 0.01 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr17_+_18117358 | 0.01 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr4_-_44509477 | 0.00 |
ENSDART00000102949
ENSDART00000135262 |
si:dkeyp-100h4.1
|
si:dkeyp-100h4.1 |
| chr12_-_18408566 | 0.00 |
ENSDART00000078767
ENSDART00000152748 |
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr2_+_44687147 | 0.00 |
ENSDART00000141605
|
yeats2
|
YEATS domain containing 2 |
| chr4_-_38829098 | 0.00 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr16_-_46558521 | 0.00 |
ENSDART00000189406
|
si:dkey-152b24.8
|
si:dkey-152b24.8 |
| chr4_-_71267461 | 0.00 |
ENSDART00000161219
ENSDART00000151838 |
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr4_+_58668661 | 0.00 |
ENSDART00000188815
|
CR388148.2
|
|
| chr24_-_25256230 | 0.00 |
ENSDART00000155780
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr4_-_34775762 | 0.00 |
ENSDART00000167262
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr9_+_42270043 | 0.00 |
ENSDART00000137435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr4_-_34776147 | 0.00 |
ENSDART00000167498
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr6_+_27418541 | 0.00 |
ENSDART00000187410
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr8_+_22405477 | 0.00 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr15_+_44133972 | 0.00 |
ENSDART00000157846
ENSDART00000191082 |
CU655961.2
|
|
| chr4_+_67994238 | 0.00 |
ENSDART00000160968
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr3_+_1749793 | 0.00 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr4_-_63559311 | 0.00 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_+_57192681 | 0.00 |
ENSDART00000190438
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr4_-_49582758 | 0.00 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr9_-_1970071 | 0.00 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr4_+_34119824 | 0.00 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr1_-_57629639 | 0.00 |
ENSDART00000158984
|
zmp:0000001289
|
zmp:0000001289 |
| chr20_+_36730049 | 0.00 |
ENSDART00000045948
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr12_+_46605600 | 0.00 |
ENSDART00000123533
|
FADS6
|
fatty acid desaturase 6 |
| chr10_+_37927100 | 0.00 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr4_-_43616331 | 0.00 |
ENSDART00000183211
ENSDART00000150240 |
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr15_+_19682013 | 0.00 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr4_+_76973771 | 0.00 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr4_+_47782411 | 0.00 |
ENSDART00000161728
|
si:ch211-196h24.2
|
si:ch211-196h24.2 |
| chr7_+_35268054 | 0.00 |
ENSDART00000113842
|
dpep2
|
dipeptidase 2 |
| chr20_+_13175379 | 0.00 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.1 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |