Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for nr6a1a
Z-value: 0.66
Transcription factors associated with nr6a1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1a
|
ENSDARG00000101508 | nuclear receptor subfamily 6, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1a | dr11_v1_chr8_-_52909850_52909850 | -0.22 | 3.8e-01 | Click! |
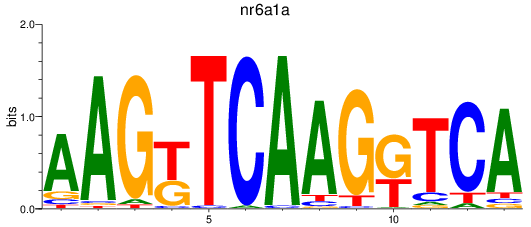
Activity profile of nr6a1a motif
Sorted Z-values of nr6a1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_29509133 | 1.99 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr3_-_3448095 | 1.95 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr2_-_50225411 | 1.87 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr24_-_9979342 | 1.68 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr24_-_9989634 | 1.62 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr21_-_43636595 | 1.56 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr16_-_17197546 | 1.56 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr18_+_8912113 | 1.52 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr5_-_37103487 | 1.51 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr12_+_30788912 | 1.32 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr7_-_41013575 | 1.29 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr11_+_45287541 | 1.26 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr17_-_40956035 | 1.13 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr14_+_35414632 | 1.11 |
ENSDART00000191516
ENSDART00000084914 |
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr10_+_44719697 | 1.05 |
ENSDART00000158087
|
scarb1
|
scavenger receptor class B, member 1 |
| chr4_+_9478500 | 1.01 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr10_-_36793412 | 1.01 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr5_-_41124241 | 0.99 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr11_+_14333441 | 0.97 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr16_-_34195002 | 0.95 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr5_+_22579975 | 0.90 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr24_+_17334682 | 0.88 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr5_+_9218318 | 0.86 |
ENSDART00000137774
|
si:ch211-12e13.1
|
si:ch211-12e13.1 |
| chr11_+_45219558 | 0.86 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr1_-_22652424 | 0.85 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr3_+_14512670 | 0.84 |
ENSDART00000161403
|
rab3db
|
RAB3D, member RAS oncogene family, b |
| chr9_-_21238159 | 0.84 |
ENSDART00000146764
ENSDART00000102143 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr22_-_22337382 | 0.83 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr22_+_25086942 | 0.82 |
ENSDART00000061117
|
rrbp1b
|
ribosome binding protein 1b |
| chr25_+_17860962 | 0.81 |
ENSDART00000163153
|
pth1a
|
parathyroid hormone 1a |
| chr21_+_35328025 | 0.81 |
ENSDART00000136211
|
rars
|
arginyl-tRNA synthetase |
| chr25_+_7492663 | 0.81 |
ENSDART00000166496
|
cat
|
catalase |
| chr1_-_8566567 | 0.79 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr9_-_12888082 | 0.78 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr2_+_32743807 | 0.78 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr22_+_25086567 | 0.77 |
ENSDART00000192114
ENSDART00000177284 ENSDART00000180296 ENSDART00000190384 |
rrbp1b
|
ribosome binding protein 1b |
| chr11_+_24314148 | 0.76 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr14_+_26224731 | 0.75 |
ENSDART00000168673
|
gm2a
|
GM2 ganglioside activator |
| chr5_-_11971946 | 0.73 |
ENSDART00000166285
|
si:ch73-47f2.1
|
si:ch73-47f2.1 |
| chr5_+_50879545 | 0.71 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr5_+_31779911 | 0.71 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr9_-_21238616 | 0.71 |
ENSDART00000191840
ENSDART00000189127 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr3_+_31662126 | 0.70 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr1_+_24557414 | 0.69 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr7_-_31618166 | 0.68 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr15_+_28410664 | 0.68 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr10_-_26218354 | 0.67 |
ENSDART00000180764
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr22_+_21152847 | 0.67 |
ENSDART00000007321
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr10_-_16868211 | 0.65 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr14_+_26224541 | 0.64 |
ENSDART00000128971
|
gm2a
|
GM2 ganglioside activator |
| chr12_+_28955766 | 0.63 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr5_-_55201172 | 0.61 |
ENSDART00000134937
|
srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr3_-_13599482 | 0.61 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr22_+_21153031 | 0.60 |
ENSDART00000166342
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr14_-_36412473 | 0.60 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr14_+_30272891 | 0.60 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr8_-_39859688 | 0.59 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr8_-_54216499 | 0.58 |
ENSDART00000193678
|
mbd4
|
methyl-CpG binding domain protein 4 |
| chr8_-_26709959 | 0.58 |
ENSDART00000135215
|
tmem51a
|
transmembrane protein 51a |
| chr10_-_25543227 | 0.58 |
ENSDART00000007778
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr25_-_35143360 | 0.56 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr9_+_24095677 | 0.55 |
ENSDART00000150443
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr23_-_545942 | 0.54 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr10_+_33393829 | 0.54 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr13_-_11763186 | 0.53 |
ENSDART00000102381
ENSDART00000187819 |
ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_+_25126629 | 0.53 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr18_+_6706140 | 0.52 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr21_-_21178410 | 0.52 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr2_+_13694450 | 0.51 |
ENSDART00000077259
ENSDART00000189485 |
ebna1bp2
|
EBNA1 binding protein 2 |
| chr21_+_1587722 | 0.50 |
ENSDART00000013581
|
wdr91
|
WD repeat domain 91 |
| chr9_-_10804796 | 0.50 |
ENSDART00000134911
|
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr5_-_42180205 | 0.50 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr21_+_19319804 | 0.50 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr24_+_19542323 | 0.48 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr16_+_50741154 | 0.48 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr24_-_10393969 | 0.48 |
ENSDART00000106260
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr23_+_12852655 | 0.48 |
ENSDART00000183015
|
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr5_+_31791001 | 0.48 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr12_-_999762 | 0.47 |
ENSDART00000127003
ENSDART00000084076 ENSDART00000152425 |
mettl9
|
methyltransferase like 9 |
| chr9_-_46276626 | 0.46 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr24_-_10828560 | 0.46 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr6_-_33924883 | 0.46 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr23_+_40139765 | 0.46 |
ENSDART00000185376
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr22_-_9861531 | 0.45 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr20_+_3277620 | 0.45 |
ENSDART00000067397
ENSDART00000135194 |
ndufaf7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr22_-_9860792 | 0.44 |
ENSDART00000155908
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr19_-_3724605 | 0.44 |
ENSDART00000123757
|
smim13
|
small integral membrane protein 13 |
| chr9_-_7287375 | 0.44 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr9_+_24088062 | 0.43 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr18_-_20608300 | 0.43 |
ENSDART00000140632
|
bcl2l13
|
BCL2 like 13 |
| chr7_+_16033923 | 0.43 |
ENSDART00000161669
ENSDART00000114062 |
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr20_-_28404362 | 0.43 |
ENSDART00000055932
ENSDART00000188161 |
pigh
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr1_+_52633367 | 0.42 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr1_+_52632856 | 0.42 |
ENSDART00000011725
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr12_+_28956374 | 0.41 |
ENSDART00000033878
|
znf668
|
zinc finger protein 668 |
| chr5_+_31959954 | 0.41 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr13_+_21677767 | 0.41 |
ENSDART00000165166
|
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr10_+_7719796 | 0.41 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr5_+_42124706 | 0.40 |
ENSDART00000020044
ENSDART00000156372 |
shpk
|
sedoheptulokinase |
| chr1_+_17527342 | 0.39 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr1_-_55118745 | 0.39 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr17_+_6538733 | 0.39 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr24_+_30521619 | 0.38 |
ENSDART00000162903
|
dpyda.3
|
dihydropyrimidine dehydrogenase a, tandem duplicate 3 |
| chr2_+_58377395 | 0.38 |
ENSDART00000193511
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr20_-_23219964 | 0.38 |
ENSDART00000144933
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr20_+_23007470 | 0.38 |
ENSDART00000134639
ENSDART00000132916 |
si:ch73-249k16.4
|
si:ch73-249k16.4 |
| chr24_+_34069675 | 0.37 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr14_-_25095808 | 0.37 |
ENSDART00000184244
|
matr3l1.1
|
matrin 3-like 1.1 |
| chr2_+_30032303 | 0.37 |
ENSDART00000151841
|
rbm33b
|
RNA binding motif protein 33b |
| chr20_-_30931139 | 0.36 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr8_-_2153147 | 0.36 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr21_-_16087857 | 0.36 |
ENSDART00000144955
ENSDART00000180621 ENSDART00000135605 |
mzt2b
|
mitotic spindle organizing protein 2B |
| chr1_+_17527931 | 0.35 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr6_-_29051773 | 0.35 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr20_+_4392687 | 0.35 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr21_+_15592426 | 0.35 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr6_-_11759860 | 0.35 |
ENSDART00000151296
|
si:ch211-162i14.1
|
si:ch211-162i14.1 |
| chr10_+_11265387 | 0.35 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr6_-_7686594 | 0.34 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr12_-_17152139 | 0.34 |
ENSDART00000152478
|
stambpl1
|
STAM binding protein-like 1 |
| chr9_-_10805231 | 0.33 |
ENSDART00000193913
ENSDART00000078348 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr12_+_28888975 | 0.33 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr11_+_15878343 | 0.33 |
ENSDART00000167191
ENSDART00000171862 ENSDART00000163992 ENSDART00000170065 |
pank4
|
pantothenate kinase 4 |
| chr13_+_33368503 | 0.33 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr15_+_32405959 | 0.32 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr15_-_30984804 | 0.32 |
ENSDART00000157005
|
nf1a
|
neurofibromin 1a |
| chr23_-_35756115 | 0.32 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr13_+_2617555 | 0.32 |
ENSDART00000162208
|
plpp4
|
phospholipid phosphatase 4 |
| chr8_-_8489886 | 0.32 |
ENSDART00000183334
|
abt1
|
activator of basal transcription 1 |
| chr24_-_25096199 | 0.31 |
ENSDART00000185076
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr6_-_42949184 | 0.31 |
ENSDART00000147208
|
edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr10_-_36618674 | 0.31 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr7_+_20383841 | 0.30 |
ENSDART00000052906
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr16_+_25184207 | 0.29 |
ENSDART00000147584
|
hcst
|
hematopoietic cell signal transducer |
| chr5_-_35252761 | 0.29 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr18_-_226800 | 0.29 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr3_+_21189766 | 0.29 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr20_-_39789846 | 0.29 |
ENSDART00000188418
|
rnf217
|
ring finger protein 217 |
| chr13_-_42724645 | 0.29 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr8_+_2878756 | 0.28 |
ENSDART00000168107
|
crb2b
|
crumbs family member 2b |
| chr24_+_5789790 | 0.28 |
ENSDART00000189600
|
BX470065.1
|
|
| chr12_-_13650344 | 0.28 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr17_-_21162821 | 0.28 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr5_-_69004007 | 0.28 |
ENSDART00000137443
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr2_-_20120904 | 0.27 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr21_+_43702016 | 0.27 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr1_+_57348756 | 0.26 |
ENSDART00000063750
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr15_+_2191900 | 0.26 |
ENSDART00000185913
|
rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr21_+_15709061 | 0.26 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
| chr10_-_43117607 | 0.26 |
ENSDART00000148293
ENSDART00000089965 |
tmem167a
|
transmembrane protein 167A |
| chr14_+_6535426 | 0.26 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr11_+_26363435 | 0.25 |
ENSDART00000088800
|
ppcs
|
phosphopantothenoylcysteine synthetase |
| chr10_+_8550435 | 0.25 |
ENSDART00000185664
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr8_+_16726386 | 0.25 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr18_+_22220656 | 0.24 |
ENSDART00000191862
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr23_-_19953089 | 0.24 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr6_+_21536131 | 0.23 |
ENSDART00000113911
ENSDART00000188472 |
micall1a
|
MICAL-like 1a |
| chr19_+_30884960 | 0.23 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr6_-_47246948 | 0.23 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr1_+_36911471 | 0.23 |
ENSDART00000148640
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr16_+_47428721 | 0.23 |
ENSDART00000180597
ENSDART00000032309 |
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr13_-_14926318 | 0.23 |
ENSDART00000142785
|
cdc25b
|
cell division cycle 25B |
| chr22_-_4407871 | 0.22 |
ENSDART00000162523
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr23_+_16633951 | 0.22 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr22_+_31207226 | 0.21 |
ENSDART00000180016
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr6_+_6491013 | 0.21 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr14_-_35414559 | 0.21 |
ENSDART00000145033
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr2_-_56649883 | 0.21 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr9_+_27379193 | 0.21 |
ENSDART00000142656
|
tlr20.4
|
toll-like receptor 20, tandem duplicate 4 |
| chr2_+_7818368 | 0.21 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr2_+_37110504 | 0.20 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr7_+_17096281 | 0.20 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr2_+_51028269 | 0.20 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr14_+_16034447 | 0.20 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr22_+_10163901 | 0.20 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr16_-_18960613 | 0.20 |
ENSDART00000183197
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr3_-_10677890 | 0.20 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr7_-_37917517 | 0.20 |
ENSDART00000173795
|
heatr3
|
HEAT repeat containing 3 |
| chr5_-_54792239 | 0.19 |
ENSDART00000056213
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_+_42338309 | 0.19 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr3_-_584950 | 0.19 |
ENSDART00000164752
|
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr4_-_30362840 | 0.19 |
ENSDART00000165929
|
znf1083
|
zinc finger protein 1083 |
| chr11_-_44621405 | 0.19 |
ENSDART00000160639
|
tbce
|
tubulin folding cofactor E |
| chr8_-_20245892 | 0.19 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr20_-_33790003 | 0.18 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr3_-_5644028 | 0.18 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr3_+_58455428 | 0.18 |
ENSDART00000074129
|
zgc:171489
|
zgc:171489 |
| chr19_+_30885258 | 0.17 |
ENSDART00000143394
|
yars
|
tyrosyl-tRNA synthetase |
| chr6_+_19383267 | 0.17 |
ENSDART00000166549
|
mchr1a
|
melanin-concentrating hormone receptor 1a |
| chr19_-_103289 | 0.17 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr4_+_77948517 | 0.17 |
ENSDART00000149305
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr21_+_20901505 | 0.17 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr19_+_5134624 | 0.17 |
ENSDART00000151324
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr7_+_19482084 | 0.17 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr13_-_49666615 | 0.17 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr11_-_97817 | 0.16 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr21_-_33126697 | 0.16 |
ENSDART00000189293
ENSDART00000084559 |
CU019662.1
|
|
| chr2_-_5199431 | 0.16 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr15_+_14854666 | 0.16 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr1_-_23110740 | 0.16 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 1.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 2.6 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.7 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 0.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 0.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.5 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 1.9 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 0.8 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.8 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 0.8 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.0 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.7 | GO:0006212 | uracil catabolic process(GO:0006212) beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) uracil metabolic process(GO:0019860) |
| 0.1 | 2.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.4 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 1.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 1.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 1.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.3 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.3 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.7 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 1.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.6 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 1.2 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.0 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.0 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 1.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.7 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.5 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.5 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 1.9 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 1.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.6 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.7 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.5 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 0.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.6 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0008106 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.2 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.3 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |