Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
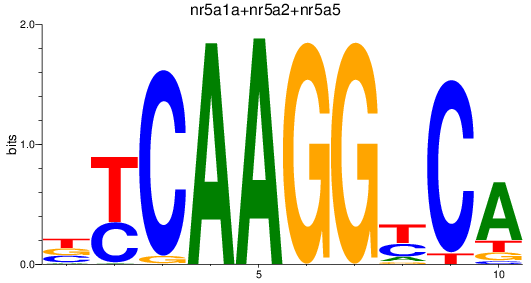
Results for nr5a1a+nr5a2+nr5a5
Z-value: 1.04
Transcription factors associated with nr5a1a+nr5a2+nr5a5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr5a5
|
ENSDARG00000039116 | nuclear receptor subfamily 5, group A, member 5 |
|
nr5a2
|
ENSDARG00000100940 | nuclear receptor subfamily 5, group A, member 2 |
|
nr5a1a
|
ENSDARG00000103176 | nuclear receptor subfamily 5, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr5a1a | dr11_v1_chr8_-_52871056_52871056 | 0.89 | 7.7e-07 | Click! |
| nr5a2 | dr11_v1_chr22_-_22719440_22719440 | 0.64 | 3.9e-03 | Click! |
| nr5a5 | dr11_v1_chr3_-_53465223_53465249 | 0.51 | 3.0e-02 | Click! |
Activity profile of nr5a1a+nr5a2+nr5a5 motif
Sorted Z-values of nr5a1a+nr5a2+nr5a5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_24488652 | 5.31 |
ENSDART00000052067
|
insl3
|
insulin-like 3 (Leydig cell) |
| chr13_+_50375800 | 4.45 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr21_+_198269 | 3.73 |
ENSDART00000193485
ENSDART00000163696 |
gsdf
|
gonadal somatic cell derived factor |
| chr15_-_20916251 | 3.40 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr7_-_59256806 | 2.98 |
ENSDART00000167955
|
m1ap
|
meiosis 1 associated protein |
| chr16_+_52512025 | 2.96 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr8_-_36554675 | 2.93 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr19_-_12145765 | 2.85 |
ENSDART00000032474
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr22_+_7439476 | 2.78 |
ENSDART00000021594
ENSDART00000063389 |
zgc:92041
|
zgc:92041 |
| chr9_+_21194445 | 2.61 |
ENSDART00000061321
|
hsd3b1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr19_-_12145390 | 2.56 |
ENSDART00000143087
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr22_-_2886937 | 2.41 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr21_+_17956856 | 2.40 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr8_-_21372446 | 2.32 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr16_-_25085327 | 2.32 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr23_+_27345826 | 2.31 |
ENSDART00000132635
|
tmprss12
|
transmembrane (C-terminal) protease, serine 12 |
| chr8_-_48770603 | 2.19 |
ENSDART00000159712
|
pimr184
|
Pim proto-oncogene, serine/threonine kinase, related 184 |
| chr5_+_20693724 | 2.18 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr2_+_17508323 | 2.17 |
ENSDART00000169703
|
pimr199
|
Pim proto-oncogene, serine/threonine kinase, related 199 |
| chr10_+_25355308 | 2.17 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr14_-_36397768 | 2.14 |
ENSDART00000185199
ENSDART00000052562 |
spata4
|
spermatogenesis associated 4 |
| chr12_+_45200744 | 1.97 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr6_-_21091948 | 1.96 |
ENSDART00000057348
|
inha
|
inhibin, alpha |
| chr22_-_20761759 | 1.93 |
ENSDART00000013803
|
amh
|
anti-Mullerian hormone |
| chr18_-_44285539 | 1.90 |
ENSDART00000137222
|
pimr179
|
Pim proto-oncogene, serine/threonine kinase, related 179 |
| chr3_-_52674089 | 1.87 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr6_+_7250824 | 1.87 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr22_+_7480465 | 1.80 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr22_-_36934040 | 1.78 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr7_+_34688527 | 1.78 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr5_-_42544522 | 1.76 |
ENSDART00000157968
|
BX510949.1
|
|
| chr8_-_29719393 | 1.76 |
ENSDART00000077635
|
si:ch211-103n10.5
|
si:ch211-103n10.5 |
| chr18_-_24996634 | 1.72 |
ENSDART00000170210
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr11_-_8226088 | 1.66 |
ENSDART00000173364
|
si:cabz01021067.1
|
si:cabz01021067.1 |
| chr25_+_3326885 | 1.66 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr3_-_1434135 | 1.65 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr11_-_8271374 | 1.63 |
ENSDART00000168253
|
pimr202
|
Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr1_+_54911458 | 1.62 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chrM_+_9735 | 1.60 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr20_+_54738210 | 1.60 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr1_+_9994811 | 1.59 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr10_-_27197044 | 1.57 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr14_+_45732081 | 1.57 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr11_+_25472758 | 1.56 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr13_-_44738574 | 1.55 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr25_+_27873836 | 1.52 |
ENSDART00000163801
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr11_+_25259058 | 1.51 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr3_+_1182315 | 1.49 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr10_-_16065185 | 1.48 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr7_-_28148310 | 1.45 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr12_-_30583668 | 1.38 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr6_-_43922813 | 1.37 |
ENSDART00000123341
|
prok2
|
prokineticin 2 |
| chr7_-_28549361 | 1.37 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr7_-_24204665 | 1.35 |
ENSDART00000167141
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr11_-_8288925 | 1.33 |
ENSDART00000168524
|
pimr202
|
Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr3_+_26081343 | 1.33 |
ENSDART00000134647
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr22_+_7439186 | 1.33 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr9_+_17348745 | 1.29 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr13_-_2215213 | 1.26 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr24_+_22021675 | 1.24 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr7_+_19904136 | 1.22 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr6_-_49078263 | 1.21 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr25_+_3327071 | 1.19 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr4_-_9764767 | 1.18 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr17_-_12385308 | 1.15 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_-_24204200 | 1.14 |
ENSDART00000087298
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr21_+_30355767 | 1.12 |
ENSDART00000189948
|
CR749164.1
|
|
| chr3_+_39568290 | 1.08 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr7_+_568819 | 1.07 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr20_+_218886 | 1.07 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr25_+_245018 | 1.07 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr12_-_48477031 | 1.05 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr19_+_619200 | 1.04 |
ENSDART00000050125
|
nupl2
|
nucleoporin like 2 |
| chr8_-_52715911 | 1.03 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr20_+_35086242 | 1.03 |
ENSDART00000114262
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr12_+_24738962 | 1.01 |
ENSDART00000105751
|
fshr
|
follicle stimulating hormone receptor |
| chr2_-_20599315 | 1.01 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr7_+_20475788 | 1.01 |
ENSDART00000171155
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr7_+_20467549 | 1.01 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr1_+_52735484 | 1.00 |
ENSDART00000182076
|
CABZ01021532.1
|
|
| chr22_+_7472420 | 1.00 |
ENSDART00000132049
|
CELA1 (1 of many)
|
si:dkey-57c15.4 |
| chr15_+_20403903 | 0.99 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr11_-_10770053 | 0.98 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr5_-_32445835 | 0.97 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr22_-_34551568 | 0.97 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr12_+_7865470 | 0.96 |
ENSDART00000161683
|
BX548028.1
|
|
| chr8_+_23213320 | 0.95 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr13_-_6218248 | 0.95 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr14_+_35892802 | 0.95 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr1_+_580642 | 0.93 |
ENSDART00000147633
|
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr5_-_36328688 | 0.93 |
ENSDART00000011399
|
efnb1
|
ephrin-B1 |
| chr18_+_14529005 | 0.92 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr19_+_24488403 | 0.92 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr14_-_5678457 | 0.88 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr21_+_3960583 | 0.88 |
ENSDART00000149788
|
setx
|
senataxin |
| chr22_+_7462997 | 0.88 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr21_-_293146 | 0.85 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr15_-_23645810 | 0.84 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr19_-_30404096 | 0.84 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr13_-_20540790 | 0.82 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr3_+_13624815 | 0.82 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr22_+_38194151 | 0.80 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr2_+_49864219 | 0.79 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr23_+_25708787 | 0.78 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr11_-_29650930 | 0.77 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr19_-_23871941 | 0.77 |
ENSDART00000134551
|
pimr92
|
Pim proto-oncogene, serine/threonine kinase, related 92 |
| chr25_+_13791627 | 0.77 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr6_+_37894914 | 0.76 |
ENSDART00000148817
|
oca2
|
oculocutaneous albinism II |
| chr23_+_36083529 | 0.76 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr6_+_25257728 | 0.75 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr7_-_38658411 | 0.75 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr21_-_26071142 | 0.73 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr4_+_11375894 | 0.72 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr22_+_7486867 | 0.72 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr14_-_48260744 | 0.71 |
ENSDART00000183955
|
rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr3_+_22442445 | 0.71 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr18_+_45504362 | 0.70 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr7_+_25913225 | 0.70 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr16_-_42013858 | 0.70 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr7_-_25895189 | 0.68 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr17_-_43287290 | 0.68 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr19_-_30403922 | 0.67 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr15_+_19544052 | 0.67 |
ENSDART00000062560
|
zgc:77784
|
zgc:77784 |
| chr15_-_21877726 | 0.67 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr22_+_15624371 | 0.66 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr15_-_2657508 | 0.66 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr5_-_38155005 | 0.66 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr17_-_6451801 | 0.66 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr21_-_5879897 | 0.65 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr11_+_40306579 | 0.64 |
ENSDART00000167504
|
si:dkey-207b20.2
|
si:dkey-207b20.2 |
| chr8_+_45003659 | 0.63 |
ENSDART00000132663
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr10_+_36184751 | 0.63 |
ENSDART00000162639
|
or108-2
|
odorant receptor, family D, subfamily 108, member 2 |
| chr20_+_10539293 | 0.63 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr11_+_705727 | 0.62 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr1_-_44940830 | 0.62 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr7_-_26087807 | 0.62 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr8_+_12930216 | 0.62 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr16_+_43347966 | 0.61 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr12_+_15622621 | 0.60 |
ENSDART00000079784
|
plcd3b
|
phospholipase C, delta 3b |
| chr14_-_31060082 | 0.60 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr2_-_55298075 | 0.60 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr8_-_17926814 | 0.60 |
ENSDART00000147344
|
lhx8b
|
LIM homeobox 8b |
| chr22_+_7742211 | 0.59 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr3_-_7170661 | 0.59 |
ENSDART00000190345
|
BX005085.4
|
|
| chr11_+_25111846 | 0.59 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr24_-_31846366 | 0.59 |
ENSDART00000155295
|
steap2
|
STEAP family member 2, metalloreductase |
| chr17_-_44584811 | 0.58 |
ENSDART00000165059
ENSDART00000165252 |
slc35f4
|
solute carrier family 35, member F4 |
| chr3_-_32859335 | 0.57 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr19_+_10831362 | 0.57 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr11_-_26180714 | 0.57 |
ENSDART00000173597
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr19_-_5103313 | 0.57 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr9_+_32930622 | 0.56 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr19_-_5103141 | 0.56 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_+_31821815 | 0.56 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr22_-_36876133 | 0.56 |
ENSDART00000147006
|
kng1
|
kininogen 1 |
| chr11_+_40208239 | 0.56 |
ENSDART00000166171
|
si:ch211-193i15.1
|
si:ch211-193i15.1 |
| chr7_+_22293894 | 0.56 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr20_+_49119633 | 0.55 |
ENSDART00000151435
|
cd109
|
CD109 molecule |
| chr11_-_18604542 | 0.55 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr9_-_1970071 | 0.54 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr22_-_17574511 | 0.54 |
ENSDART00000181496
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr2_-_50198328 | 0.54 |
ENSDART00000074511
ENSDART00000137704 |
ahrrb
|
aryl-hydrocarbon receptor repressor b |
| chr13_+_42124566 | 0.54 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr25_-_13842618 | 0.52 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr2_-_58201173 | 0.52 |
ENSDART00000166282
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr25_+_36316280 | 0.51 |
ENSDART00000152735
|
si:ch211-113a14.11
|
si:ch211-113a14.11 |
| chr9_+_12934536 | 0.51 |
ENSDART00000134484
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr5_-_22027357 | 0.51 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr11_-_843811 | 0.51 |
ENSDART00000173331
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr21_+_26071874 | 0.50 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr5_-_63109232 | 0.50 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr11_+_807153 | 0.50 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr19_-_23768865 | 0.49 |
ENSDART00000147081
|
pimr87
|
Pim proto-oncogene, serine/threonine kinase, related 87 |
| chr15_+_23788154 | 0.49 |
ENSDART00000181482
|
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr20_+_10545514 | 0.49 |
ENSDART00000153667
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr10_+_42374770 | 0.48 |
ENSDART00000020000
|
COX5B
|
zgc:86599 |
| chr24_+_26432541 | 0.48 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr5_+_38276582 | 0.48 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr6_-_8480815 | 0.47 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr2_-_11468654 | 0.46 |
ENSDART00000131229
|
slc44a5a
|
solute carrier family 44, member 5a |
| chr5_+_25072894 | 0.46 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr6_+_8176486 | 0.46 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr22_+_7497319 | 0.45 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr17_-_8862424 | 0.45 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr7_+_61480296 | 0.45 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr12_-_3940768 | 0.45 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr23_+_24604481 | 0.45 |
ENSDART00000146332
|
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr20_-_9760424 | 0.44 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr7_-_28696556 | 0.44 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr14_+_6159356 | 0.44 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr15_+_1004680 | 0.44 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr4_+_77021784 | 0.44 |
ENSDART00000135345
ENSDART00000133855 |
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr10_+_466926 | 0.44 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr14_-_40389699 | 0.43 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr22_-_26865361 | 0.43 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr6_+_58832155 | 0.43 |
ENSDART00000144842
|
dctn2
|
dynactin 2 (p50) |
| chr23_-_1056808 | 0.43 |
ENSDART00000081961
|
zgc:113423
|
zgc:113423 |
| chr6_-_1514767 | 0.42 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr11_-_25213651 | 0.42 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr9_-_23891102 | 0.42 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr5a1a+nr5a2+nr5a5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.6 | 3.0 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.5 | 6.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 1.3 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 0.8 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 0.9 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.2 | 0.6 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.2 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 2.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 1.0 | GO:0008584 | male gonad development(GO:0008584) |
| 0.2 | 1.0 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 1.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 0.6 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.7 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.9 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.5 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.4 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.3 | GO:1901295 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.4 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 2.1 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 2.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 2.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.5 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 1.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.6 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 5.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.4 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.2 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.1 | 0.4 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.7 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.2 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 2.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.8 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.2 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 1.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 1.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) long-chain fatty acid import(GO:0044539) |
| 0.0 | 2.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.4 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 1.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 12.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 1.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 1.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.5 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 1.0 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.5 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.1 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 1.7 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.1 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.4 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 1.3 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.3 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 2.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 3.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.5 | 3.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 1.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.3 | 2.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 0.9 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 2.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 1.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 0.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 7.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 0.7 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 2.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 0.6 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.5 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.4 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 1.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.7 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 7.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 5.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 1.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 5.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.6 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0016641 | FMN binding(GO:0010181) oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 7.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 2.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 1.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 6.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |