Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
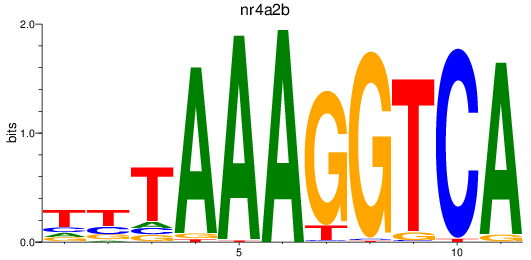
Results for nr4a2b
Z-value: 0.41
Transcription factors associated with nr4a2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a2b
|
ENSDARG00000044532 | nuclear receptor subfamily 4, group A, member 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a2b | dr11_v1_chr6_+_12462079_12462079 | -0.24 | 3.4e-01 | Click! |
Activity profile of nr4a2b motif
Sorted Z-values of nr4a2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_5332784 | 0.75 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr22_+_30184039 | 0.59 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr17_+_26753967 | 0.58 |
ENSDART00000025096
|
larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr8_+_13106760 | 0.54 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr21_-_41369370 | 0.49 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr22_+_29113796 | 0.43 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr8_-_22542467 | 0.42 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr7_+_22792132 | 0.41 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr21_-_41369539 | 0.41 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr22_-_11833317 | 0.39 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr18_+_27515640 | 0.37 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr3_+_33745014 | 0.35 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr11_-_44999858 | 0.34 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr6_-_41135215 | 0.34 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr7_+_30202104 | 0.33 |
ENSDART00000173525
|
wdr76
|
WD repeat domain 76 |
| chr5_-_13076779 | 0.32 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr13_+_27298673 | 0.30 |
ENSDART00000142922
|
slc17a5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr1_-_23110740 | 0.29 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr14_+_30413312 | 0.29 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr7_-_42206720 | 0.27 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr10_+_11265387 | 0.27 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr5_+_6670945 | 0.27 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr24_-_27256338 | 0.27 |
ENSDART00000105768
|
zmynd11
|
zinc finger, MYND-type containing 11 |
| chr24_-_27256673 | 0.26 |
ENSDART00000181182
|
zmynd11
|
zinc finger, MYND-type containing 11 |
| chr2_-_37312927 | 0.26 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr1_-_40102836 | 0.26 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr11_-_31226578 | 0.24 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr13_-_24260609 | 0.24 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr15_-_16946124 | 0.24 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr2_+_16585549 | 0.23 |
ENSDART00000134135
|
si:dkeyp-13a3.10
|
si:dkeyp-13a3.10 |
| chr17_-_15189397 | 0.23 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr21_-_36571804 | 0.23 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr24_-_8214253 | 0.22 |
ENSDART00000160432
|
CABZ01077956.1
|
|
| chr3_-_54500354 | 0.22 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr6_-_31348999 | 0.22 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_-_10082244 | 0.21 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr11_+_5681762 | 0.21 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr3_-_45250924 | 0.21 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr5_-_68093169 | 0.21 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr14_+_30413758 | 0.20 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr12_-_850457 | 0.20 |
ENSDART00000022688
|
tob1b
|
transducer of ERBB2, 1b |
| chr8_-_8698607 | 0.19 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr5_+_24156170 | 0.19 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr7_+_38667066 | 0.19 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr4_+_11723852 | 0.18 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr23_-_45504991 | 0.18 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr23_+_30730121 | 0.18 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr8_-_49756171 | 0.18 |
ENSDART00000140609
|
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr8_+_53120278 | 0.18 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr14_-_26482096 | 0.18 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr3_+_17806213 | 0.17 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr11_-_38492564 | 0.17 |
ENSDART00000102850
|
elk4
|
ELK4, ETS-domain protein |
| chr13_+_7241170 | 0.16 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr9_+_24065855 | 0.16 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr5_-_5669879 | 0.16 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr1_-_58868306 | 0.15 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr15_+_33691455 | 0.15 |
ENSDART00000170329
|
hdac12
|
histone deacetylase 12 |
| chr24_+_22022109 | 0.14 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr4_-_2975461 | 0.14 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_+_42206847 | 0.13 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr5_-_10007897 | 0.13 |
ENSDART00000109052
|
CR936408.1
|
Danio rerio uncharacterized LOC799523 (LOC799523), mRNA. |
| chr2_+_34767171 | 0.13 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr24_+_684924 | 0.12 |
ENSDART00000186103
|
si:ch211-188f17.1
|
si:ch211-188f17.1 |
| chr13_-_27660955 | 0.12 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr5_-_41831646 | 0.12 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr14_+_30340251 | 0.11 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr13_-_41155472 | 0.11 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr25_+_16214854 | 0.10 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr15_+_31526225 | 0.10 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr2_-_38282079 | 0.10 |
ENSDART00000145808
|
rnf212b
|
si:ch211-10e2.1 |
| chr22_+_11153590 | 0.10 |
ENSDART00000188207
|
bcor
|
BCL6 corepressor |
| chr5_+_60928576 | 0.10 |
ENSDART00000131041
|
doc2b
|
double C2-like domains, beta |
| chr5_+_36611128 | 0.09 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr5_+_37406358 | 0.09 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr7_-_24046999 | 0.09 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr23_+_21544227 | 0.09 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr24_+_26997798 | 0.09 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr1_-_20593778 | 0.08 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr11_-_40647190 | 0.08 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr4_-_8060962 | 0.08 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr25_+_22017182 | 0.08 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr18_-_17513426 | 0.08 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr6_+_13206516 | 0.07 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr7_+_25913225 | 0.07 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr19_+_17358850 | 0.07 |
ENSDART00000123519
|
ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr18_-_42333428 | 0.07 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr7_+_34620418 | 0.07 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr18_-_4957022 | 0.07 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr25_-_16969922 | 0.07 |
ENSDART00000111158
|
tigara
|
tp53-induced glycolysis and apoptosis regulator a |
| chr15_-_16884912 | 0.06 |
ENSDART00000062135
|
zgc:103681
|
zgc:103681 |
| chr16_-_25680666 | 0.06 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr3_-_25377163 | 0.06 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr12_+_27156943 | 0.06 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr9_+_31795343 | 0.06 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr9_-_21067971 | 0.06 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr4_-_18775548 | 0.06 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr17_+_30545895 | 0.06 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr9_-_20853439 | 0.06 |
ENSDART00000028247
ENSDART00000133321 |
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr15_+_20403903 | 0.06 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr23_-_24488696 | 0.06 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr6_+_3282809 | 0.06 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr1_-_30689004 | 0.06 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr14_+_35023923 | 0.05 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr24_-_1021318 | 0.05 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr7_+_34296789 | 0.05 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr9_+_35860975 | 0.05 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr10_-_25561751 | 0.05 |
ENSDART00000147089
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr3_+_24190207 | 0.05 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr20_-_26937453 | 0.05 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr10_+_9561066 | 0.04 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr12_+_14084291 | 0.04 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr11_-_21363834 | 0.04 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr7_-_31794878 | 0.04 |
ENSDART00000099748
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr12_+_30368145 | 0.04 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr15_-_11341635 | 0.04 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr7_-_31794476 | 0.03 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr1_-_21287724 | 0.03 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr6_-_10320676 | 0.03 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr8_-_979735 | 0.03 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr17_-_1705013 | 0.03 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr1_+_44826367 | 0.02 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr5_+_37087583 | 0.02 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr6_-_39764995 | 0.02 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr5_-_67721368 | 0.02 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr7_-_19146925 | 0.02 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr24_+_39211288 | 0.02 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr18_+_27926839 | 0.02 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr6_-_39765546 | 0.02 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr6_-_8498908 | 0.02 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr16_-_39195318 | 0.02 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr23_+_24272421 | 0.02 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr5_-_63218919 | 0.01 |
ENSDART00000149979
|
tecta
|
tectorin alpha |
| chr8_-_44004135 | 0.01 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr5_-_33959868 | 0.01 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr5_-_41838354 | 0.01 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr7_+_31871830 | 0.01 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr1_+_19332837 | 0.01 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr10_+_41668483 | 0.01 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr8_-_42238543 | 0.01 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr16_-_11859309 | 0.01 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr15_-_21239416 | 0.01 |
ENSDART00000155787
|
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr16_-_16761164 | 0.01 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr11_+_1409622 | 0.01 |
ENSDART00000157042
|
ppp1r3db
|
protein phosphatase 1, regulatory subunit 3Db |
| chr3_-_22212764 | 0.01 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr1_+_44826593 | 0.00 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr18_-_2433011 | 0.00 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr21_-_22724980 | 0.00 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr8_+_20825987 | 0.00 |
ENSDART00000133309
|
si:ch211-133l5.4
|
si:ch211-133l5.4 |
| chr20_+_35086242 | 0.00 |
ENSDART00000114262
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr5_-_3839285 | 0.00 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr10_-_29892486 | 0.00 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr6_-_21189295 | 0.00 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr10_+_7563755 | 0.00 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr3_-_46817838 | 0.00 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr25_-_31898552 | 0.00 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.2 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0051196 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.0 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |