Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
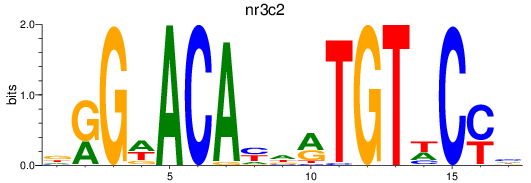
Results for nr3c2
Z-value: 0.23
Transcription factors associated with nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c2
|
ENSDARG00000102082 | nuclear receptor subfamily 3, group C, member 2 |
|
nr3c2
|
ENSDARG00000115513 | nuclear receptor subfamily 3, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr3c2 | dr11_v1_chr1_-_37087966_37087966 | -0.86 | 4.5e-06 | Click! |
Activity profile of nr3c2 motif
Sorted Z-values of nr3c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_40051425 | 0.79 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr14_-_46113321 | 0.62 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr8_+_23034718 | 0.53 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr6_+_4255319 | 0.51 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr2_-_32512648 | 0.44 |
ENSDART00000170674
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr10_+_20364009 | 0.43 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr7_-_51749683 | 0.42 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr13_-_33700461 | 0.40 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr5_+_30518441 | 0.36 |
ENSDART00000132664
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr11_-_43473824 | 0.32 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr15_+_40188076 | 0.30 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr22_-_34979139 | 0.28 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr18_-_20608025 | 0.27 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2 like 13 |
| chr17_-_6613458 | 0.26 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr18_-_20608300 | 0.23 |
ENSDART00000140632
|
bcl2l13
|
BCL2 like 13 |
| chr15_+_23951560 | 0.23 |
ENSDART00000191133
|
myo18ab
|
myosin XVIIIAb |
| chr24_-_20641000 | 0.19 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr10_+_8554929 | 0.19 |
ENSDART00000190849
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr3_+_13850163 | 0.18 |
ENSDART00000164872
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr25_+_10416583 | 0.15 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr11_+_34522554 | 0.15 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr20_+_54336137 | 0.15 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr10_+_29849497 | 0.15 |
ENSDART00000099994
ENSDART00000132212 |
hspa8
|
heat shock protein 8 |
| chr18_-_19491162 | 0.15 |
ENSDART00000090413
|
snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr1_-_28473350 | 0.14 |
ENSDART00000190608
ENSDART00000148175 |
si:ch1073-440b2.1
|
si:ch1073-440b2.1 |
| chr8_+_39767915 | 0.14 |
ENSDART00000017153
|
hps4
|
Hermansky-Pudlak syndrome 4 |
| chr24_-_23784701 | 0.12 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr5_-_54792239 | 0.12 |
ENSDART00000056213
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr11_+_34523132 | 0.10 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr5_-_9540641 | 0.10 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr10_-_24689725 | 0.10 |
ENSDART00000079566
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr25_-_13408760 | 0.10 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr9_+_22388686 | 0.10 |
ENSDART00000182731
ENSDART00000181462 |
dgkg
|
diacylglycerol kinase, gamma |
| chr17_+_6276559 | 0.09 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr24_-_38097305 | 0.09 |
ENSDART00000124321
|
crp2
|
C-reactive protein 2 |
| chr15_-_18429550 | 0.07 |
ENSDART00000136208
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr11_+_5880562 | 0.07 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr15_-_24883956 | 0.07 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr3_-_25275364 | 0.04 |
ENSDART00000163782
ENSDART00000145420 ENSDART00000133718 ENSDART00000055492 |
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr22_-_16275236 | 0.04 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr2_+_29257942 | 0.02 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr22_+_19538626 | 0.02 |
ENSDART00000189174
|
BX936298.2
|
|
| chr18_-_44611252 | 0.02 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr19_+_7916790 | 0.02 |
ENSDART00000081584
|
si:dkey-266f7.1
|
si:dkey-266f7.1 |
| chr3_-_37476475 | 0.02 |
ENSDART00000148107
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr21_+_6291027 | 0.01 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr11_-_34147205 | 0.01 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr15_+_31462664 | 0.01 |
ENSDART00000174267
ENSDART00000185665 ENSDART00000060087 |
or102-1
|
odorant receptor, family C, subfamily 102, member 1 |
| chr25_+_28772632 | 0.00 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.8 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.5 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.6 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.1 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |