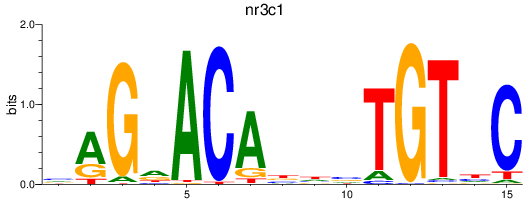
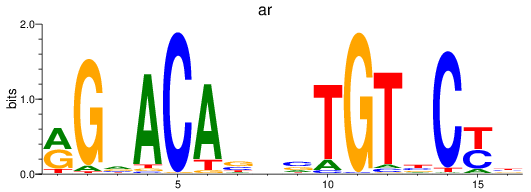
Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for nr3c1_ar
Z-value: 0.45
Transcription factors associated with nr3c1_ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c1
|
ENSDARG00000025032 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000112480 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000116957 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
ar
|
ENSDARG00000067976 | androgen receptor |
|
ar
|
ENSDARG00000114287 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr3c1 | dr11_v1_chr14_-_23801389_23801389 | 0.74 | 4.2e-04 | Click! |
| ar | dr11_v1_chr5_+_35561607_35561607 | 0.73 | 6.6e-04 | Click! |
Activity profile of nr3c1_ar motif
Sorted Z-values of nr3c1_ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_33589667 | 1.96 |
ENSDART00000152097
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr3_-_33422738 | 1.19 |
ENSDART00000075493
|
ccdc103
|
coiled-coil domain containing 103 |
| chr3_+_58833306 | 0.92 |
ENSDART00000113223
|
igl1c3
|
immunoglobulin light 1 constant 3 |
| chr10_+_26667475 | 0.91 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr16_-_45225520 | 0.86 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr8_-_50525360 | 0.78 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr18_-_24996634 | 0.69 |
ENSDART00000170210
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr1_+_57311901 | 0.69 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr11_+_2642013 | 0.66 |
ENSDART00000111324
|
zgc:193807
|
zgc:193807 |
| chr25_+_3358701 | 0.66 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr13_-_37619159 | 0.64 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr6_+_41096058 | 0.63 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr4_+_12358822 | 0.60 |
ENSDART00000172557
ENSDART00000150627 |
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr6_+_41099787 | 0.51 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr20_+_35445462 | 0.51 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr13_+_39236141 | 0.51 |
ENSDART00000111458
|
zgc:172136
|
zgc:172136 |
| chr8_-_13315304 | 0.49 |
ENSDART00000142596
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr16_-_22225295 | 0.48 |
ENSDART00000163519
|
LO017682.1
|
|
| chr15_-_5742531 | 0.48 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr16_-_12266153 | 0.48 |
ENSDART00000113345
|
leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr12_+_49135755 | 0.48 |
ENSDART00000153460
|
si:zfos-911d5.4
|
si:zfos-911d5.4 |
| chr20_+_25340814 | 0.47 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr18_+_16744307 | 0.44 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr8_-_13315567 | 0.44 |
ENSDART00000132685
ENSDART00000168635 |
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr16_+_50089417 | 0.43 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr25_-_6420800 | 0.42 |
ENSDART00000153768
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr25_-_5189989 | 0.42 |
ENSDART00000097520
|
shisal1b
|
shisa like 1b |
| chr4_-_56068511 | 0.41 |
ENSDART00000168345
|
znf1133
|
zinc finger protein 1133 |
| chr4_+_11723852 | 0.41 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr7_+_58699718 | 0.40 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr20_-_6812688 | 0.39 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr20_-_32752457 | 0.38 |
ENSDART00000153287
|
si:dkey-6f10.3
|
si:dkey-6f10.3 |
| chr10_-_15672862 | 0.37 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr8_-_11202378 | 0.36 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr7_-_16598212 | 0.35 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr10_+_26669177 | 0.35 |
ENSDART00000143402
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr18_-_44316920 | 0.35 |
ENSDART00000098599
|
si:ch211-151h10.2
|
si:ch211-151h10.2 |
| chr24_-_20956781 | 0.33 |
ENSDART00000142080
|
kpna1
|
karyopherin alpha 1 (importin alpha 5) |
| chr14_+_39258569 | 0.32 |
ENSDART00000103298
|
diaph2
|
diaphanous-related formin 2 |
| chr1_-_59176949 | 0.32 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr25_-_3469576 | 0.32 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr21_-_19316985 | 0.31 |
ENSDART00000141596
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr2_-_20788698 | 0.31 |
ENSDART00000181823
|
CABZ01020455.1
|
|
| chr22_-_36519590 | 0.30 |
ENSDART00000129318
|
CABZ01045212.1
|
|
| chr8_+_35172594 | 0.30 |
ENSDART00000177146
|
BX897670.1
|
|
| chr16_+_18535618 | 0.29 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr7_-_29271738 | 0.29 |
ENSDART00000109030
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr9_+_23009608 | 0.29 |
ENSDART00000079879
|
si:dkey-91i10.3
|
si:dkey-91i10.3 |
| chr14_+_29769336 | 0.28 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr5_-_69523816 | 0.28 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr10_+_29431529 | 0.28 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr15_+_37559570 | 0.28 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr4_-_789645 | 0.28 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr25_-_32449235 | 0.28 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr16_+_10776688 | 0.27 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr19_+_5418006 | 0.27 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr4_-_38507880 | 0.27 |
ENSDART00000183225
|
si:ch211-209n20.3
|
si:ch211-209n20.3 |
| chr4_-_15420452 | 0.27 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr6_+_40354424 | 0.27 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr17_+_33767890 | 0.26 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr21_+_43402794 | 0.26 |
ENSDART00000185572
|
frmd7
|
FERM domain containing 7 |
| chr23_+_32039386 | 0.26 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr22_+_37874691 | 0.26 |
ENSDART00000028565
|
ahsg1
|
alpha-2-HS-glycoprotein 1 |
| chr5_-_41494831 | 0.26 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr21_+_32339158 | 0.26 |
ENSDART00000161723
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr7_+_52178109 | 0.26 |
ENSDART00000174099
|
katnb1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr5_+_37649206 | 0.26 |
ENSDART00000149151
ENSDART00000097723 |
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr25_-_3470910 | 0.25 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr6_-_41091151 | 0.25 |
ENSDART00000154963
ENSDART00000153818 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr11_-_236766 | 0.25 |
ENSDART00000163978
|
dusp7
|
dual specificity phosphatase 7 |
| chr16_+_25126935 | 0.25 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr4_-_12725513 | 0.25 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr23_+_35708730 | 0.24 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr7_+_6969909 | 0.24 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr14_+_52571134 | 0.24 |
ENSDART00000166708
|
rpl26
|
ribosomal protein L26 |
| chr6_-_43817761 | 0.24 |
ENSDART00000183945
|
foxp1b
|
forkhead box P1b |
| chr7_+_21331688 | 0.24 |
ENSDART00000128014
|
dnah2
|
dynein, axonemal, heavy chain 2 |
| chr22_+_38301365 | 0.23 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr21_+_6290566 | 0.23 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr20_-_791788 | 0.23 |
ENSDART00000134128
|
impg1a
|
interphotoreceptor matrix proteoglycan 1a |
| chr14_-_36863432 | 0.22 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr20_-_29475172 | 0.22 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr10_-_41906609 | 0.22 |
ENSDART00000102530
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr21_-_32462856 | 0.22 |
ENSDART00000147318
|
zgc:123105
|
zgc:123105 |
| chr16_+_26863414 | 0.22 |
ENSDART00000140673
|
galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr18_+_21273749 | 0.22 |
ENSDART00000143265
|
hydin
|
HYDIN, axonemal central pair apparatus protein |
| chr18_-_48745517 | 0.22 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr4_-_9764767 | 0.22 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr25_+_31912153 | 0.22 |
ENSDART00000153968
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr23_+_9088191 | 0.21 |
ENSDART00000030811
|
cables2b
|
Cdk5 and Abl enzyme substrate 2b |
| chr17_+_45737992 | 0.21 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr5_+_66220601 | 0.20 |
ENSDART00000129903
|
mfsd10
|
major facilitator superfamily domain containing 10 |
| chr4_+_4803698 | 0.20 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr20_+_37844035 | 0.20 |
ENSDART00000041397
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr24_-_24271629 | 0.20 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr7_+_48288762 | 0.20 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr9_+_12934536 | 0.20 |
ENSDART00000134484
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr20_-_38836161 | 0.20 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr4_+_15968483 | 0.20 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr1_+_7517454 | 0.20 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr7_-_18168493 | 0.19 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr4_+_76735113 | 0.19 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr24_+_35564668 | 0.19 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr4_-_71110826 | 0.19 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr18_+_26422124 | 0.19 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr14_-_46228094 | 0.19 |
ENSDART00000172788
|
si:ch211-113d11.5
|
si:ch211-113d11.5 |
| chr22_-_1079773 | 0.19 |
ENSDART00000136668
|
si:ch1073-15f12.3
|
si:ch1073-15f12.3 |
| chr13_+_13821054 | 0.18 |
ENSDART00000127417
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr7_-_35036770 | 0.18 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr1_+_31674297 | 0.18 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr23_+_6752828 | 0.18 |
ENSDART00000105179
|
zgc:158254
|
zgc:158254 |
| chr14_-_28001986 | 0.18 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr24_+_18714212 | 0.18 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr1_+_41898919 | 0.18 |
ENSDART00000140845
|
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr11_-_236984 | 0.18 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr11_+_34522554 | 0.17 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr15_-_3282220 | 0.17 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr12_+_32073660 | 0.17 |
ENSDART00000153245
ENSDART00000153268 |
stxbp4
|
syntaxin binding protein 4 |
| chr1_-_28885919 | 0.17 |
ENSDART00000152182
|
poglut1
|
protein O-glucosyltransferase 1 |
| chr21_-_32467099 | 0.17 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr20_+_46586678 | 0.17 |
ENSDART00000014166
ENSDART00000179266 |
jdp2b
|
Jun dimerization protein 2b |
| chr21_+_11503212 | 0.17 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr18_-_15911394 | 0.17 |
ENSDART00000091339
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr1_-_53907092 | 0.16 |
ENSDART00000007732
|
capn9
|
calpain 9 |
| chr14_+_45732081 | 0.16 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr20_+_34543365 | 0.16 |
ENSDART00000152073
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr6_+_50451337 | 0.16 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr16_+_10777116 | 0.16 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr12_-_46959990 | 0.16 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr3_-_7147226 | 0.16 |
ENSDART00000178155
|
BX005085.5
|
|
| chr4_-_69230711 | 0.16 |
ENSDART00000163977
|
si:dkey-82i20.5
|
si:dkey-82i20.5 |
| chr20_+_40769265 | 0.16 |
ENSDART00000061168
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr13_-_45523026 | 0.16 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr23_-_18415872 | 0.15 |
ENSDART00000135430
|
fam120c
|
family with sequence similarity 120C |
| chr25_+_7423770 | 0.15 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr15_-_23342752 | 0.15 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr20_+_33904258 | 0.15 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr25_+_19149241 | 0.15 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr1_-_59116617 | 0.15 |
ENSDART00000137471
ENSDART00000140490 |
MFAP4 (1 of many)
|
si:zfos-2330d3.7 |
| chr7_-_65114067 | 0.15 |
ENSDART00000110614
ENSDART00000098277 |
tmem41b
|
transmembrane protein 41B |
| chr2_-_22688651 | 0.15 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr22_-_30973791 | 0.15 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr23_+_33777519 | 0.15 |
ENSDART00000159642
|
CR381531.2
|
|
| chr11_-_42472941 | 0.15 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr5_-_43935460 | 0.15 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr22_-_11714104 | 0.14 |
ENSDART00000145265
ENSDART00000063127 ENSDART00000183743 |
crygs1
|
crystallin, gamma S1 |
| chr25_-_21154510 | 0.14 |
ENSDART00000183858
ENSDART00000086631 |
wnk1a
|
WNK lysine deficient protein kinase 1a |
| chr21_+_30563115 | 0.14 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr21_+_6291027 | 0.14 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr18_+_45114392 | 0.14 |
ENSDART00000172328
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr22_+_26813917 | 0.14 |
ENSDART00000147719
|
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr20_-_4883673 | 0.14 |
ENSDART00000145540
ENSDART00000053877 |
zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr12_-_3453589 | 0.14 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr21_-_26114886 | 0.14 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr10_+_41765944 | 0.14 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr9_-_21918963 | 0.14 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr20_-_39391833 | 0.14 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr22_+_10010292 | 0.14 |
ENSDART00000180096
|
BX324216.4
|
|
| chr8_+_53452681 | 0.14 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr5_-_5669879 | 0.13 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr19_+_19412692 | 0.13 |
ENSDART00000113580
|
wu:fc38h03
|
wu:fc38h03 |
| chr2_-_48196092 | 0.13 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr6_-_22064158 | 0.13 |
ENSDART00000153585
|
cfap100
|
cilia and flagella associated protein 100 |
| chr6_+_9130989 | 0.13 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr18_-_2639351 | 0.13 |
ENSDART00000168106
|
relt
|
RELT, TNF receptor |
| chr16_-_19026733 | 0.13 |
ENSDART00000104826
|
pimr98
|
Pim proto-oncogene, serine/threonine kinase, related 98 |
| chr3_-_5067585 | 0.13 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr21_+_5802377 | 0.13 |
ENSDART00000151316
|
ccng2
|
cyclin G2 |
| chr3_+_20012891 | 0.13 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr22_+_28995391 | 0.13 |
ENSDART00000164486
|
pimr99
|
Pim proto-oncogene, serine/threonine kinase, related 99 |
| chr24_-_37472727 | 0.13 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr21_+_45502773 | 0.12 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr13_-_37636147 | 0.12 |
ENSDART00000144065
|
si:dkey-188i13.8
|
si:dkey-188i13.8 |
| chr13_+_21826369 | 0.12 |
ENSDART00000165150
ENSDART00000192115 |
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr11_+_34523132 | 0.12 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr13_+_29510023 | 0.12 |
ENSDART00000187398
|
chst3a
|
carbohydrate (chondroitin 6) sulfotransferase 3a |
| chr3_-_54669185 | 0.12 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr13_+_22295905 | 0.12 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr11_+_13630107 | 0.12 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr3_+_62205858 | 0.12 |
ENSDART00000126807
|
zgc:173575
|
zgc:173575 |
| chr10_+_22891126 | 0.12 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr7_+_49272702 | 0.12 |
ENSDART00000083389
|
abtb2a
|
ankyrin repeat and BTB (POZ) domain containing 2a |
| chr21_-_32289356 | 0.12 |
ENSDART00000183050
|
clk4b
|
CDC-like kinase 4b |
| chr21_+_25054420 | 0.12 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr25_-_12803723 | 0.12 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr24_+_23742690 | 0.11 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr16_+_46430627 | 0.11 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr10_-_24343507 | 0.11 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr6_-_13114821 | 0.10 |
ENSDART00000164579
ENSDART00000165375 |
zgc:194469
|
zgc:194469 |
| chr5_+_9360394 | 0.10 |
ENSDART00000124642
|
FP236810.2
|
|
| chr20_+_7084154 | 0.10 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr15_-_21669618 | 0.10 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr5_+_23256187 | 0.10 |
ENSDART00000168717
ENSDART00000142915 |
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr19_-_10730488 | 0.10 |
ENSDART00000126033
|
slc9a3.1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 1 |
| chr25_-_24046870 | 0.10 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr5_-_12093618 | 0.10 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr9_-_41818760 | 0.10 |
ENSDART00000140601
|
si:dkeyp-30e7.2
|
si:dkeyp-30e7.2 |
| chr11_+_45343245 | 0.10 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr17_-_45387134 | 0.10 |
ENSDART00000010975
|
tmem206
|
transmembrane protein 206 |
| chr24_+_13316737 | 0.10 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c1_ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 2.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.3 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.2 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.1 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.7 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.0 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.0 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0006595 | polyamine metabolic process(GO:0006595) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.3 | GO:0008352 | katanin complex(GO:0008352) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.2 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |