Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
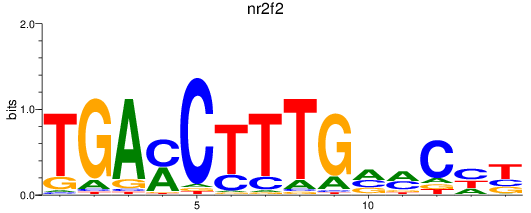
Results for nr2f2
Z-value: 1.29
Transcription factors associated with nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f2
|
ENSDARG00000040926 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f2 | dr11_v1_chr18_-_23874929_23874929 | -0.80 | 6.4e-05 | Click! |
Activity profile of nr2f2 motif
Sorted Z-values of nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_27564458 | 8.73 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr10_-_21542702 | 6.97 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr10_+_19569052 | 6.27 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr19_-_27564980 | 5.43 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr1_+_24387659 | 5.22 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr17_-_2573021 | 5.20 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2595736 | 5.20 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2590222 | 5.17 |
ENSDART00000185711
|
CR759892.1
|
|
| chr17_-_2578026 | 5.05 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr23_+_2740741 | 4.82 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr20_-_43741159 | 4.28 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr9_+_8380728 | 3.45 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr9_+_33216945 | 3.39 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr17_-_2584423 | 3.36 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr7_-_24520866 | 3.34 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr18_-_43866526 | 2.64 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr12_-_10508952 | 2.42 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr22_-_24992532 | 2.40 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr19_-_3056235 | 1.99 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr22_-_7050 | 1.87 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr15_-_30857350 | 1.86 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr9_-_41153896 | 1.78 |
ENSDART00000059667
|
wdr75
|
WD repeat domain 75 |
| chr11_+_41838801 | 1.72 |
ENSDART00000014871
|
akr7a3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr18_-_43866001 | 1.69 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr7_-_20611039 | 1.65 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr3_-_36419641 | 1.63 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr7_+_24520518 | 1.52 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr9_+_22782027 | 1.49 |
ENSDART00000090816
|
rif1
|
replication timing regulatory factor 1 |
| chr20_+_36806398 | 1.49 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr16_-_39131666 | 1.44 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr5_+_9218318 | 1.44 |
ENSDART00000137774
|
si:ch211-12e13.1
|
si:ch211-12e13.1 |
| chr18_+_1615 | 1.43 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr8_+_21159122 | 1.41 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr1_-_53468160 | 1.41 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr16_-_25233515 | 1.40 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr6_+_40952031 | 1.36 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr25_-_35143360 | 1.36 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr7_-_51775688 | 1.32 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr10_+_43039947 | 1.31 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr7_+_24114694 | 1.31 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr3_+_31662126 | 1.25 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr23_+_30730121 | 1.23 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr7_+_24115082 | 1.21 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr18_-_127558 | 1.19 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr13_+_8696825 | 1.15 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr18_+_36664654 | 1.14 |
ENSDART00000128707
ENSDART00000098972 |
capn12
|
calpain 12 |
| chr3_-_34561624 | 1.13 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr3_-_5829501 | 1.12 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr24_-_21172122 | 1.10 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr4_-_19693978 | 1.09 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr17_-_49407091 | 1.05 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr14_+_36414856 | 1.04 |
ENSDART00000123343
ENSDART00000015761 |
neil3
|
nei-like DNA glycosylase 3 |
| chr17_-_6613458 | 1.00 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr15_+_8394819 | 0.99 |
ENSDART00000147086
|
tmem160
|
transmembrane protein 160 |
| chr18_-_46183462 | 0.99 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr24_-_27409599 | 0.99 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr2_-_20120904 | 0.97 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr25_-_24202576 | 0.95 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr18_-_158541 | 0.92 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr14_-_32876280 | 0.92 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr3_+_34121156 | 0.91 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr2_-_389867 | 0.90 |
ENSDART00000004848
|
wrnip1
|
Werner helicase interacting protein 1 |
| chr6_-_31682135 | 0.89 |
ENSDART00000153988
|
cachd1
|
cache domain containing 1 |
| chr20_-_51656512 | 0.88 |
ENSDART00000129965
|
LO018154.1
|
|
| chr21_-_9446747 | 0.88 |
ENSDART00000158790
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr3_+_17933132 | 0.86 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr10_+_35417099 | 0.85 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr7_-_58244220 | 0.85 |
ENSDART00000180450
|
unm_hu7910
|
un-named hu7910 |
| chr9_-_25255490 | 0.85 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr16_-_25680666 | 0.83 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr7_-_20464133 | 0.81 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr10_+_15340768 | 0.81 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr2_-_56649883 | 0.79 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr6_-_11759860 | 0.79 |
ENSDART00000151296
|
si:ch211-162i14.1
|
si:ch211-162i14.1 |
| chr5_+_43470544 | 0.78 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr10_+_45128375 | 0.78 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr7_-_62003831 | 0.78 |
ENSDART00000113585
ENSDART00000062704 |
plaa
|
phospholipase A2-activating protein |
| chr10_-_22797959 | 0.77 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr3_+_12755535 | 0.74 |
ENSDART00000161286
|
cyp2k17
|
cytochrome P450, family 2, subfamily K, polypeptide17 |
| chr1_+_1712140 | 0.74 |
ENSDART00000081047
|
atp1a1a.1
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 1 |
| chr4_-_9579299 | 0.73 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr14_+_30413312 | 0.72 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr10_+_24690534 | 0.72 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr4_-_858434 | 0.72 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr6_-_28117995 | 0.72 |
ENSDART00000147253
|
si:ch73-194h10.3
|
si:ch73-194h10.3 |
| chr17_-_6076266 | 0.72 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr25_-_20049449 | 0.71 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr2_-_38312757 | 0.71 |
ENSDART00000167274
|
ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr16_-_19568388 | 0.70 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr10_+_1849874 | 0.70 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr19_-_11031145 | 0.70 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr3_+_34180835 | 0.67 |
ENSDART00000055252
|
timm29
|
translocase of inner mitochondrial membrane 29 |
| chr13_+_39277178 | 0.66 |
ENSDART00000113259
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr17_-_6076084 | 0.65 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr23_-_18595020 | 0.65 |
ENSDART00000187032
ENSDART00000191047 |
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr20_+_3277620 | 0.65 |
ENSDART00000067397
ENSDART00000135194 |
ndufaf7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr2_+_58377395 | 0.65 |
ENSDART00000193511
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr3_+_39566999 | 0.65 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr25_+_36348401 | 0.64 |
ENSDART00000103006
|
hist1h2a3
|
histone cluster 1 H2A family member 3 |
| chr10_+_20364009 | 0.64 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr3_+_26288981 | 0.62 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr9_+_41690153 | 0.61 |
ENSDART00000100226
|
ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr11_-_127785 | 0.61 |
ENSDART00000162658
|
ARFGAP1
|
ADP ribosylation factor GTPase activating protein 1 |
| chr24_-_23716097 | 0.61 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr2_-_3614005 | 0.59 |
ENSDART00000110399
|
pter
|
phosphotriesterase related |
| chr9_+_54695981 | 0.59 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr16_+_4078240 | 0.58 |
ENSDART00000160890
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr17_-_22137199 | 0.57 |
ENSDART00000089689
|
mrps5
|
mitochondrial ribosomal protein S5 |
| chr8_-_8349653 | 0.57 |
ENSDART00000025214
|
tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr6_-_1587291 | 0.57 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr16_+_33143503 | 0.57 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr14_+_23874062 | 0.56 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr15_+_40074923 | 0.56 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr12_+_1609563 | 0.56 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr23_-_44786844 | 0.56 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr22_-_31517300 | 0.55 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr14_-_30876299 | 0.55 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr7_-_19600181 | 0.55 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr25_+_22017182 | 0.55 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr12_+_6195191 | 0.55 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr9_-_28990649 | 0.54 |
ENSDART00000078823
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr2_+_9821757 | 0.54 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr22_+_2417105 | 0.53 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr9_+_21151138 | 0.52 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr25_+_19008497 | 0.52 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr16_-_27149202 | 0.51 |
ENSDART00000179726
|
tmem245
|
transmembrane protein 245 |
| chr7_+_24496894 | 0.51 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr23_-_18567088 | 0.50 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr3_+_24050043 | 0.50 |
ENSDART00000151788
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr9_-_4606463 | 0.48 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr5_-_61588998 | 0.48 |
ENSDART00000050912
|
pex12
|
peroxisomal biogenesis factor 12 |
| chr16_+_7991274 | 0.48 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr16_-_32672883 | 0.48 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr17_+_26718648 | 0.48 |
ENSDART00000154492
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr2_-_41571454 | 0.47 |
ENSDART00000022643
|
znf622
|
zinc finger protein 622 |
| chr18_+_27515640 | 0.47 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr15_+_47618221 | 0.47 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr12_+_48972915 | 0.46 |
ENSDART00000170695
|
lrit1b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1b |
| chr21_+_41743493 | 0.46 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr8_-_22542467 | 0.46 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr8_-_20243389 | 0.45 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr4_+_1619584 | 0.45 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr3_+_31680592 | 0.45 |
ENSDART00000172456
|
mylk5
|
myosin, light chain kinase 5 |
| chr21_-_1640547 | 0.45 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr17_+_48314724 | 0.45 |
ENSDART00000125617
|
smoc1
|
SPARC related modular calcium binding 1 |
| chr16_-_19568795 | 0.44 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr18_-_12957451 | 0.43 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr23_+_9522942 | 0.43 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr9_+_17429170 | 0.43 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr9_-_9960940 | 0.43 |
ENSDART00000092164
|
prmt2
|
protein arginine methyltransferase 2 |
| chr3_+_40289418 | 0.43 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr13_+_24552254 | 0.43 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr24_+_39137001 | 0.43 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr1_-_57172294 | 0.42 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr2_+_5948534 | 0.42 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr6_+_55819038 | 0.42 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr16_-_21692024 | 0.41 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr17_+_15534815 | 0.40 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr21_+_7188943 | 0.40 |
ENSDART00000172174
|
agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr22_+_30502555 | 0.40 |
ENSDART00000139128
ENSDART00000104747 |
zgc:171679
|
zgc:171679 |
| chr22_-_11833317 | 0.39 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr20_-_27733683 | 0.39 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr7_+_65398161 | 0.39 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr17_+_15213496 | 0.39 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr13_-_42724645 | 0.39 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr5_+_63340637 | 0.39 |
ENSDART00000143742
|
si:ch73-376l24.4
|
si:ch73-376l24.4 |
| chr4_+_25917915 | 0.39 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr17_+_33418475 | 0.39 |
ENSDART00000169145
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr11_-_5563498 | 0.38 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr13_-_33114933 | 0.38 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr5_+_4016271 | 0.37 |
ENSDART00000113627
ENSDART00000105832 ENSDART00000121415 |
ggnbp2
|
gametogenetin binding protein 2 |
| chr23_+_9522781 | 0.37 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr6_-_14004772 | 0.37 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr5_+_63322093 | 0.35 |
ENSDART00000158086
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr2_+_30531726 | 0.35 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr10_-_11385155 | 0.35 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr11_-_44999858 | 0.34 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr13_-_41546779 | 0.34 |
ENSDART00000163331
|
pcdh15a
|
protocadherin-related 15a |
| chr4_-_75172216 | 0.34 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr13_-_9311253 | 0.34 |
ENSDART00000058056
|
mrps26
|
mitochondrial ribosomal protein S26 |
| chr9_+_54984537 | 0.33 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr14_+_22680485 | 0.33 |
ENSDART00000167829
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr20_-_9658405 | 0.33 |
ENSDART00000023809
|
nid2b
|
nidogen 2b (osteonidogen) |
| chr15_+_17345609 | 0.33 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr18_+_25546227 | 0.32 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr14_+_22152060 | 0.32 |
ENSDART00000082697
ENSDART00000054982 |
gabra6a
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6a |
| chr18_+_31410652 | 0.31 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr22_-_10774735 | 0.31 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr14_+_989733 | 0.31 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr1_-_19215336 | 0.31 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr12_+_30368145 | 0.31 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr22_-_26595027 | 0.31 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr5_-_32396929 | 0.30 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr12_+_26670778 | 0.29 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr19_-_340347 | 0.29 |
ENSDART00000139924
|
golph3l
|
golgi phosphoprotein 3-like |
| chr19_+_48024457 | 0.28 |
ENSDART00000163823
|
kpnb1
|
karyopherin (importin) beta 1 |
| chr13_-_3516473 | 0.28 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr23_-_46126444 | 0.28 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr4_-_36139585 | 0.27 |
ENSDART00000132071
|
znf992
|
zinc finger protein 992 |
| chr11_+_6115621 | 0.27 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 1.1 | 18.8 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.7 | 2.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.6 | 4.8 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.5 | 5.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.4 | 1.1 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.4 | 1.4 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.3 | 1.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 1.0 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 1.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.4 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 0.7 | GO:1905067 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.2 | 0.5 | GO:0090151 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.2 | 0.9 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.6 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.4 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.5 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.7 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.6 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.9 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 2.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.0 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 2.4 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.4 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.0 | 0.7 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.9 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 1.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 1.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.7 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 1.5 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 2.0 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.7 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.3 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 1.5 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.7 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.9 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.9 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.0 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.8 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 1.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.0 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 1.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0048798 | somatic muscle development(GO:0007525) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 0.8 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 0.7 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 1.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 2.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 1.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.5 | GO:0098799 | mitochondrial sorting and assembly machinery complex(GO:0001401) outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 1.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 11.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 5.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 18.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 1.4 | 4.3 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 1.3 | 5.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.5 | 1.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.4 | 1.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.3 | 0.9 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.2 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 0.9 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 2.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.0 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.2 | 1.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.2 | 0.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 2.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.2 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.1 | 0.9 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 20.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 1.0 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.2 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.6 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 1.0 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.9 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 2.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 1.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.6 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.0 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 2.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 1.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |