Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
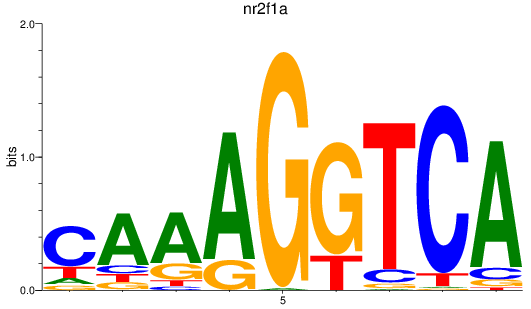
Results for nr2f1a
Z-value: 0.58
Transcription factors associated with nr2f1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f1a
|
ENSDARG00000052695 | nuclear receptor subfamily 2, group F, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f1a | dr11_v1_chr5_+_49744713_49744713 | -0.46 | 5.7e-02 | Click! |
Activity profile of nr2f1a motif
Sorted Z-values of nr2f1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_27564458 | 2.15 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr6_+_154556 | 2.09 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr1_+_24387659 | 1.98 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr10_-_21542702 | 1.76 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr17_-_2590222 | 1.48 |
ENSDART00000185711
|
CR759892.1
|
|
| chr3_-_3448095 | 1.46 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr17_-_2573021 | 1.42 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_-_27564980 | 1.33 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr17_-_2595736 | 1.27 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr10_+_19569052 | 1.16 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr17_-_2578026 | 1.11 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_26820698 | 1.09 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr11_+_18037729 | 1.03 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr11_+_18130300 | 1.03 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr19_-_27570333 | 1.00 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr11_+_18053333 | 0.99 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr3_+_3454610 | 0.98 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr11_+_18157260 | 0.96 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr7_-_24520866 | 0.90 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr8_-_23780334 | 0.87 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr16_-_17200120 | 0.85 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr3_-_5829501 | 0.82 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr22_-_7050 | 0.82 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr10_+_24692076 | 0.76 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr16_+_53603945 | 0.75 |
ENSDART00000083513
|
sacm1lb
|
SAC1 like phosphatidylinositide phosphatase b |
| chr11_+_45287541 | 0.75 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr16_-_24668620 | 0.71 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr9_-_32300611 | 0.70 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr2_+_30249977 | 0.65 |
ENSDART00000109160
ENSDART00000135171 |
tmem70
|
transmembrane protein 70 |
| chr9_-_32300783 | 0.65 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr11_-_34219211 | 0.63 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr5_-_69934558 | 0.63 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr7_+_24520518 | 0.59 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr8_+_36509885 | 0.58 |
ENSDART00000109530
|
slc7a4
|
solute carrier family 7, member 4 |
| chr16_-_39131666 | 0.57 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr10_-_21362071 | 0.57 |
ENSDART00000125167
|
avd
|
avidin |
| chr23_+_2740741 | 0.57 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr22_-_26834043 | 0.56 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr24_-_39518599 | 0.56 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr1_+_31573225 | 0.55 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr12_+_23866368 | 0.55 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr5_+_69868911 | 0.53 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr15_+_8394819 | 0.52 |
ENSDART00000147086
|
tmem160
|
transmembrane protein 160 |
| chr9_-_1639884 | 0.52 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr7_-_51775688 | 0.52 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr21_+_37411547 | 0.51 |
ENSDART00000076320
|
mrps17
|
mitochondrial ribosomal protein S17 |
| chr14_-_14659023 | 0.51 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr3_+_39566999 | 0.50 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr20_+_36806398 | 0.50 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr13_+_22480496 | 0.50 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr2_-_56649883 | 0.49 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr4_-_4535189 | 0.48 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr2_+_16710889 | 0.48 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr19_-_8798178 | 0.48 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr23_-_27050083 | 0.46 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr24_-_27409599 | 0.45 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr3_-_36419641 | 0.45 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr25_-_35143360 | 0.45 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr18_-_43866526 | 0.44 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr3_+_7771420 | 0.44 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr24_-_10393969 | 0.44 |
ENSDART00000106260
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr4_-_965267 | 0.44 |
ENSDART00000093289
|
atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr3_+_39568290 | 0.44 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr15_+_20799943 | 0.43 |
ENSDART00000154665
|
aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr25_+_22571475 | 0.43 |
ENSDART00000161559
|
stra6
|
stimulated by retinoic acid 6 |
| chr11_+_13528437 | 0.42 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr9_-_23242684 | 0.42 |
ENSDART00000053282
ENSDART00000179770 |
ccnt2a
|
cyclin T2a |
| chr12_+_47439 | 0.42 |
ENSDART00000114732
|
mrpl58
|
mitochondrial ribosomal protein L58 |
| chr18_-_43866001 | 0.42 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr23_-_22072716 | 0.41 |
ENSDART00000054362
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr5_-_57879138 | 0.41 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr23_+_39346774 | 0.40 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr16_+_25608778 | 0.40 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr12_-_10508952 | 0.40 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr18_+_35229115 | 0.40 |
ENSDART00000129624
ENSDART00000184596 |
tbrg1
|
transforming growth factor beta regulator 1 |
| chr6_+_40952031 | 0.38 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_+_31662126 | 0.38 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr15_-_30857350 | 0.37 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr22_+_39084829 | 0.37 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr18_-_127558 | 0.37 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr1_-_9249943 | 0.37 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr12_+_46634736 | 0.37 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr19_-_3056235 | 0.37 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr8_+_2530065 | 0.36 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr23_+_39346930 | 0.36 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr21_-_28640316 | 0.36 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr11_-_24063196 | 0.36 |
ENSDART00000036513
|
trib3
|
tribbles pseudokinase 3 |
| chr24_-_2843107 | 0.35 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr14_-_33478963 | 0.35 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr23_+_34030550 | 0.35 |
ENSDART00000003296
|
sars
|
seryl-tRNA synthetase |
| chr24_-_6024466 | 0.35 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr19_-_867071 | 0.35 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr14_-_48103207 | 0.35 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr10_+_10972795 | 0.35 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr19_-_10330778 | 0.35 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr19_+_2877079 | 0.34 |
ENSDART00000160847
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr17_+_17764979 | 0.34 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr3_-_13599482 | 0.34 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr25_-_9805269 | 0.34 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr24_+_16393302 | 0.34 |
ENSDART00000188670
ENSDART00000081759 ENSDART00000177790 |
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr3_+_27770110 | 0.34 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr15_+_26933196 | 0.33 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr12_-_45238759 | 0.33 |
ENSDART00000154859
|
trim65
|
tripartite motif containing 65 |
| chr8_+_25733872 | 0.33 |
ENSDART00000156340
|
si:ch211-167b20.8
|
si:ch211-167b20.8 |
| chr5_+_9218318 | 0.33 |
ENSDART00000137774
|
si:ch211-12e13.1
|
si:ch211-12e13.1 |
| chr12_-_28799642 | 0.33 |
ENSDART00000066303
|
mrpl10
|
mitochondrial ribosomal protein L10 |
| chr18_+_45571378 | 0.33 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr7_+_24496894 | 0.33 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr7_+_14005111 | 0.33 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr12_+_21525496 | 0.32 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr5_+_19712011 | 0.32 |
ENSDART00000131924
|
fam222a
|
family with sequence similarity 222, member A |
| chr23_+_12545114 | 0.32 |
ENSDART00000105283
ENSDART00000166990 |
si:zfos-452g4.1
|
si:zfos-452g4.1 |
| chr8_-_40183197 | 0.32 |
ENSDART00000005118
|
gpx8
|
glutathione peroxidase 8 (putative) |
| chr20_-_14941046 | 0.32 |
ENSDART00000152775
ENSDART00000152516 ENSDART00000021394 |
mettl13
|
methyltransferase like 13 |
| chr8_+_21159122 | 0.32 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr5_-_69180587 | 0.31 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr9_-_44905867 | 0.31 |
ENSDART00000138316
ENSDART00000131252 ENSDART00000179383 ENSDART00000159337 |
zgc:66484
|
zgc:66484 |
| chr20_-_23254876 | 0.31 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr10_+_43039947 | 0.31 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr18_+_44769027 | 0.31 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr25_-_35107791 | 0.30 |
ENSDART00000192819
|
zgc:165555
|
zgc:165555 |
| chr22_+_10163901 | 0.30 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr15_+_32405959 | 0.30 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr17_-_49407091 | 0.30 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr2_+_38892631 | 0.30 |
ENSDART00000134040
|
si:ch211-119o8.4
|
si:ch211-119o8.4 |
| chr8_-_45835056 | 0.30 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr5_-_69180227 | 0.30 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr8_-_2529878 | 0.29 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr1_-_57172294 | 0.29 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr10_-_8197049 | 0.29 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr19_-_42503143 | 0.29 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr18_+_20468157 | 0.29 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr2_-_20715094 | 0.29 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr3_+_18795570 | 0.29 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr14_+_1719367 | 0.29 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr16_-_25233515 | 0.28 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr17_+_15213496 | 0.28 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr6_-_7020162 | 0.28 |
ENSDART00000148982
|
bin1b
|
bridging integrator 1b |
| chr13_+_22479988 | 0.28 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr5_+_62988916 | 0.28 |
ENSDART00000123243
|
TIMM22
|
translocase of inner mitochondrial membrane 22 |
| chr13_-_31544365 | 0.28 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr1_+_24557414 | 0.28 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr9_+_24209665 | 0.27 |
ENSDART00000090280
|
zgc:153901
|
zgc:153901 |
| chr18_-_46183462 | 0.27 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr7_+_41421998 | 0.27 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr3_-_32590164 | 0.27 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr2_-_41562868 | 0.26 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr10_+_8875195 | 0.26 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr13_+_42676202 | 0.26 |
ENSDART00000099782
|
mrpl14
|
mitochondrial ribosomal protein L14 |
| chr9_+_38883388 | 0.26 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr23_+_42396934 | 0.26 |
ENSDART00000169821
|
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr12_+_10115964 | 0.26 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr1_+_45002971 | 0.26 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr11_-_30364847 | 0.26 |
ENSDART00000078387
|
pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr7_-_19600181 | 0.25 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr23_+_30730121 | 0.25 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr12_-_48943467 | 0.25 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr17_+_6538733 | 0.25 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr25_+_19008497 | 0.25 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr3_+_46764022 | 0.25 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr16_+_20871021 | 0.25 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr8_-_17997845 | 0.25 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr3_-_18189283 | 0.25 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr9_+_17429170 | 0.25 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr9_+_54686686 | 0.25 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr8_-_17167819 | 0.24 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr3_+_23029484 | 0.24 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr21_+_5080789 | 0.24 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr6_-_2627488 | 0.24 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr7_-_8738827 | 0.24 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr24_-_32522587 | 0.24 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr15_-_6946286 | 0.23 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr12_+_34378733 | 0.23 |
ENSDART00000018939
|
tha1
|
threonine aldolase 1 |
| chr24_-_21172122 | 0.23 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr18_+_40364732 | 0.23 |
ENSDART00000123661
ENSDART00000130847 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr12_-_54375 | 0.23 |
ENSDART00000152304
|
si:ch1073-357b18.4
|
si:ch1073-357b18.4 |
| chr6_-_39080630 | 0.23 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr19_-_48349002 | 0.23 |
ENSDART00000168468
|
si:ch73-359m17.8
|
si:ch73-359m17.8 |
| chr9_+_54695981 | 0.23 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr14_+_29200772 | 0.22 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr2_-_5728843 | 0.22 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr21_-_22681534 | 0.22 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr2_+_44977889 | 0.22 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr12_+_34827808 | 0.22 |
ENSDART00000105533
|
tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr7_-_62003831 | 0.22 |
ENSDART00000113585
ENSDART00000062704 |
plaa
|
phospholipase A2-activating protein |
| chr21_-_1972236 | 0.22 |
ENSDART00000192858
ENSDART00000189962 ENSDART00000182461 ENSDART00000165547 |
WDR7
|
WD repeat domain 7 |
| chr23_-_35756115 | 0.22 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr13_-_11679486 | 0.22 |
ENSDART00000108694
|
si:ch211-195e19.1
|
si:ch211-195e19.1 |
| chr7_-_20611039 | 0.22 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr12_+_27589607 | 0.22 |
ENSDART00000066288
|
spata20
|
spermatogenesis associated 20 |
| chr22_-_9861531 | 0.21 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr7_+_13988075 | 0.21 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr16_-_39195318 | 0.21 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr21_-_5879897 | 0.21 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr25_+_35663933 | 0.21 |
ENSDART00000154979
|
slc2a13b
|
solute carrier family 2 (facilitated glucose transporter), member 13b |
| chr25_+_35889102 | 0.21 |
ENSDART00000023453
ENSDART00000125821 ENSDART00000135441 |
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr18_+_44768829 | 0.21 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr8_-_28339151 | 0.20 |
ENSDART00000148765
ENSDART00000149173 ENSDART00000150113 |
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr1_+_38153944 | 0.20 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr23_-_18595020 | 0.20 |
ENSDART00000187032
ENSDART00000191047 |
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 1.3 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 0.9 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.3 | 0.8 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 3.8 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 0.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.6 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.4 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.5 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.5 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.4 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.3 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.4 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.3 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.4 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 0.3 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.6 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.4 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.2 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.3 | GO:0035461 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) vitamin transmembrane transport(GO:0035461) coenzyme transport(GO:0051182) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.1 | 0.3 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0042823 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 2.0 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 1.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.2 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.4 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 2.3 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.1 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.0 | GO:0048210 | endothelial cell chemotaxis(GO:0035767) Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.0 | 0.0 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.0 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.2 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.4 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.4 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 1.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 1.3 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.3 | 3.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 0.9 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.3 | 0.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 0.9 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 0.5 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.1 | 0.4 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.5 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.5 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.2 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.5 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 1.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 4.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.8 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |