Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for nr2e1
Z-value: 0.90
Transcription factors associated with nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e1
|
ENSDARG00000017107 | nuclear receptor subfamily 2, group E, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2e1 | dr11_v1_chr20_-_32446406_32446406 | 0.80 | 6.6e-05 | Click! |
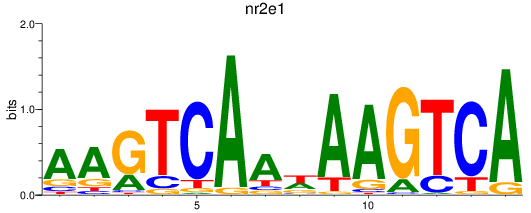
Activity profile of nr2e1 motif
Sorted Z-values of nr2e1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_14272083 | 2.93 |
ENSDART00000169986
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr12_+_18533198 | 2.54 |
ENSDART00000189729
|
meiob
|
meiosis specific with OB domains |
| chr25_-_12515741 | 2.44 |
ENSDART00000159487
|
si:ch211-280a23.2
|
si:ch211-280a23.2 |
| chr2_-_14271874 | 2.38 |
ENSDART00000189390
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr23_+_3731375 | 2.36 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr18_+_31016379 | 2.35 |
ENSDART00000172461
ENSDART00000163471 |
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr18_-_41232297 | 2.28 |
ENSDART00000036928
|
fbxo36a
|
F-box protein 36a |
| chr7_-_30367650 | 2.27 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr14_-_858985 | 2.25 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr4_-_73709502 | 2.21 |
ENSDART00000170308
|
BX855614.3
|
|
| chr16_-_35392926 | 2.07 |
ENSDART00000164482
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr6_-_31364475 | 1.96 |
ENSDART00000145715
ENSDART00000134370 |
ak4
|
adenylate kinase 4 |
| chr17_-_12408109 | 1.95 |
ENSDART00000155509
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr12_-_23128746 | 1.89 |
ENSDART00000170018
|
armc4
|
armadillo repeat containing 4 |
| chr25_-_16160494 | 1.87 |
ENSDART00000146880
|
si:dkey-80c24.4
|
si:dkey-80c24.4 |
| chr7_+_73332486 | 1.86 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr1_+_44003654 | 1.64 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr23_+_44497701 | 1.63 |
ENSDART00000149903
|
si:ch73-375g18.1
|
si:ch73-375g18.1 |
| chr10_+_10351685 | 1.62 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr16_-_39178979 | 1.43 |
ENSDART00000158894
|
si:dkey-99n20.6
|
si:dkey-99n20.6 |
| chr24_+_35827766 | 1.43 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr25_-_15507523 | 1.40 |
ENSDART00000134166
|
si:dkeyp-67e1.3
|
si:dkeyp-67e1.3 |
| chr18_-_37407235 | 1.39 |
ENSDART00000132315
|
cep126
|
centrosomal protein 126 |
| chr1_+_44336897 | 1.39 |
ENSDART00000157578
|
si:ch211-165a10.1
|
si:ch211-165a10.1 |
| chr1_+_7546259 | 1.36 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr7_+_26029672 | 1.30 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr1_+_57311901 | 1.28 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr15_-_39948635 | 1.25 |
ENSDART00000114836
|
msh5
|
mutS homolog 5 |
| chr15_-_1822548 | 1.24 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr20_-_46554440 | 1.19 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr13_-_12660318 | 1.19 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr17_+_21760032 | 1.19 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr17_+_14784275 | 1.16 |
ENSDART00000005355
|
rtraf
|
RNA transcription, translation and transport factor |
| chr1_-_46244523 | 1.10 |
ENSDART00000143908
|
si:ch211-138g9.3
|
si:ch211-138g9.3 |
| chr15_-_1822166 | 1.10 |
ENSDART00000156883
|
mmp28
|
matrix metallopeptidase 28 |
| chr22_+_28969071 | 1.10 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr20_+_30725778 | 1.10 |
ENSDART00000062532
|
nhsl1b
|
NHS-like 1b |
| chr2_+_17524278 | 1.09 |
ENSDART00000165633
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr24_+_23742690 | 1.08 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr6_-_52723901 | 1.08 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr18_+_3530769 | 1.07 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr21_-_25801956 | 1.07 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr5_-_48285756 | 1.03 |
ENSDART00000183495
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr19_+_29798064 | 1.02 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr20_-_1176631 | 0.98 |
ENSDART00000152255
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr9_+_3388099 | 0.95 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr12_+_7491690 | 0.95 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr19_+_9533008 | 0.91 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr5_+_30680804 | 0.90 |
ENSDART00000078049
|
pdzd3b
|
PDZ domain containing 3b |
| chr1_-_9130136 | 0.88 |
ENSDART00000113013
|
palb2
|
partner and localizer of BRCA2 |
| chr1_+_12009673 | 0.84 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr7_-_29571615 | 0.81 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr20_+_33294428 | 0.80 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr17_+_12408188 | 0.79 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr9_-_6380653 | 0.73 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr14_-_40389699 | 0.72 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr3_-_62393449 | 0.72 |
ENSDART00000101870
ENSDART00000140782 ENSDART00000181704 |
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr1_+_14348399 | 0.71 |
ENSDART00000064468
|
crmp1
|
collapsin response mediator protein 1 |
| chr2_+_21486529 | 0.70 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr3_-_26017592 | 0.70 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr14_-_45604667 | 0.70 |
ENSDART00000167016
|
si:ch211-276i12.9
|
si:ch211-276i12.9 |
| chr15_-_4485828 | 0.66 |
ENSDART00000062868
|
tfdp2
|
transcription factor Dp-2 |
| chr7_-_41693004 | 0.65 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr3_-_26017831 | 0.64 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr20_+_30578163 | 0.61 |
ENSDART00000159485
|
fgfr1op
|
FGFR1 oncogene partner |
| chr4_-_8060962 | 0.56 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr14_-_17575764 | 0.55 |
ENSDART00000123145
|
rnf4
|
ring finger protein 4 |
| chr24_+_26895748 | 0.54 |
ENSDART00000089351
|
nceh1b.1
|
neutral cholesterol ester hydrolase 1b, tandem duplicate 1 |
| chr3_-_36606884 | 0.54 |
ENSDART00000172003
|
si:dkeyp-72e1.6
|
si:dkeyp-72e1.6 |
| chr8_+_4838114 | 0.54 |
ENSDART00000146667
|
es1
|
es1 protein |
| chr12_+_7865470 | 0.53 |
ENSDART00000161683
|
BX548028.1
|
|
| chr23_-_16485190 | 0.50 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr19_-_46775562 | 0.49 |
ENSDART00000187144
|
abrab
|
actin binding Rho activating protein b |
| chr20_+_38257766 | 0.48 |
ENSDART00000147485
ENSDART00000149160 |
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr4_-_2868112 | 0.48 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr15_-_33834577 | 0.48 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr5_-_45877387 | 0.48 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr6_+_3828560 | 0.47 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr20_+_11731039 | 0.45 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr17_+_43843536 | 0.45 |
ENSDART00000164097
|
FO704777.1
|
|
| chr16_+_20915319 | 0.43 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr23_-_19682971 | 0.43 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr7_+_13382852 | 0.42 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr12_+_9551667 | 0.42 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr7_-_3693536 | 0.42 |
ENSDART00000126381
|
si:ch211-282j17.13
|
si:ch211-282j17.13 |
| chr1_-_38538284 | 0.42 |
ENSDART00000009777
|
glra3
|
glycine receptor, alpha 3 |
| chr9_+_51147380 | 0.38 |
ENSDART00000003913
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr9_+_51265283 | 0.38 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr14_-_26377044 | 0.38 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr25_+_12640211 | 0.38 |
ENSDART00000165108
|
jph3
|
junctophilin 3 |
| chr18_-_5542351 | 0.38 |
ENSDART00000129459
|
spata5l1
|
spermatogenesis associated 5-like 1 |
| chr4_+_40953320 | 0.37 |
ENSDART00000151912
|
znf1136
|
zinc finger protein 1136 |
| chr16_-_26855936 | 0.37 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr17_-_48743076 | 0.36 |
ENSDART00000173117
|
kcnk17
|
potassium channel, subfamily K, member 17 |
| chr5_-_38197080 | 0.36 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr10_-_20669635 | 0.35 |
ENSDART00000131361
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr7_+_57108823 | 0.35 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr4_-_71983381 | 0.34 |
ENSDART00000169130
|
ftr64
|
finTRIM family, member 64 |
| chr1_+_41465771 | 0.34 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr2_+_10771787 | 0.33 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr17_+_29345606 | 0.32 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr15_+_42599501 | 0.31 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr6_-_29007493 | 0.31 |
ENSDART00000065139
|
gfi1ab
|
growth factor independent 1A transcription repressor b |
| chr19_-_38539670 | 0.31 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr19_-_27196090 | 0.29 |
ENSDART00000045616
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr7_-_70665965 | 0.29 |
ENSDART00000097710
|
ppargc1a
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr22_+_24646339 | 0.29 |
ENSDART00000183865
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr5_-_19861766 | 0.29 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr5_+_9246018 | 0.28 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr17_+_12408405 | 0.28 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr18_+_44631789 | 0.28 |
ENSDART00000144271
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr10_-_15644904 | 0.28 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr15_+_37545855 | 0.27 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr5_+_13870340 | 0.27 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr15_+_15314189 | 0.26 |
ENSDART00000156830
|
camkk1b
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha b |
| chr2_-_30249746 | 0.25 |
ENSDART00000124153
ENSDART00000142575 |
elocb
|
elongin C paralog b |
| chr1_+_41466011 | 0.25 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr10_-_20650302 | 0.25 |
ENSDART00000142028
ENSDART00000064662 |
rassf2b
|
Ras association (RalGDS/AF-6) domain family member 2b |
| chr24_+_20658942 | 0.24 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr19_+_28284120 | 0.23 |
ENSDART00000103862
|
mrpl36
|
mitochondrial ribosomal protein L36 |
| chr19_+_48176745 | 0.23 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr10_-_1625080 | 0.22 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr17_+_28073062 | 0.22 |
ENSDART00000182021
|
AL844186.1
|
|
| chr13_-_15986871 | 0.22 |
ENSDART00000189394
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr20_+_27020201 | 0.22 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr9_-_23944470 | 0.21 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr13_+_8958438 | 0.21 |
ENSDART00000165721
ENSDART00000169398 |
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr15_-_16098531 | 0.21 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr23_+_2669 | 0.20 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr2_-_38141997 | 0.20 |
ENSDART00000143469
|
trim110
|
tripartite motif containing 110 |
| chr21_+_37411547 | 0.20 |
ENSDART00000076320
|
mrps17
|
mitochondrial ribosomal protein S17 |
| chr8_-_50287949 | 0.20 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr1_+_14073891 | 0.19 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr21_-_22547496 | 0.19 |
ENSDART00000166835
ENSDART00000089030 |
myo5b
|
myosin VB |
| chr20_+_51006362 | 0.18 |
ENSDART00000028084
|
mpp5b
|
membrane protein, palmitoylated 5b (MAGUK p55 subfamily member 5) |
| chr25_-_31433512 | 0.18 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr21_-_28901095 | 0.18 |
ENSDART00000180820
|
cxxc5a
|
CXXC finger protein 5a |
| chr24_+_27511734 | 0.18 |
ENSDART00000105771
|
cxcl32b.1
|
chemokine (C-X-C motif) ligand 32b, duplicate 1 |
| chr4_+_43392655 | 0.18 |
ENSDART00000150797
|
znf1073
|
zinc finger protein 1073 |
| chr19_-_3886991 | 0.18 |
ENSDART00000162085
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr24_-_7699356 | 0.17 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr5_-_39474235 | 0.17 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr16_+_38360002 | 0.16 |
ENSDART00000087346
ENSDART00000148101 |
zgc:113232
|
zgc:113232 |
| chr5_-_50781623 | 0.15 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr15_+_37546391 | 0.15 |
ENSDART00000186625
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr16_+_20947439 | 0.15 |
ENSDART00000137344
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr14_+_44774089 | 0.14 |
ENSDART00000098643
|
tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr4_+_59748607 | 0.14 |
ENSDART00000108499
|
znf1068
|
zinc finger protein 1068 |
| chr17_+_5985933 | 0.12 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr5_-_63663811 | 0.10 |
ENSDART00000111403
|
ciz1b
|
cdkn1a interacting zinc finger protein 1b |
| chr2_-_55861351 | 0.10 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr16_+_28932038 | 0.10 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr24_+_20658760 | 0.10 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr10_-_11203164 | 0.09 |
ENSDART00000040308
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr11_-_18601955 | 0.08 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr22_-_22301672 | 0.08 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr25_-_16724913 | 0.08 |
ENSDART00000157075
|
galnt8a.2
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 2 |
| chr22_+_2409175 | 0.07 |
ENSDART00000141776
|
zgc:113220
|
zgc:113220 |
| chr4_+_60381608 | 0.07 |
ENSDART00000167351
|
znf1130
|
zinc finger protein 1130 |
| chr9_-_31135901 | 0.06 |
ENSDART00000113027
ENSDART00000193210 ENSDART00000128896 |
si:ch211-184m13.4
|
si:ch211-184m13.4 |
| chr11_-_29946640 | 0.05 |
ENSDART00000079175
|
zgc:113276
|
zgc:113276 |
| chr25_-_16742438 | 0.05 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr22_+_13977772 | 0.05 |
ENSDART00000080313
|
arl4ca
|
ADP-ribosylation factor-like 4Ca |
| chr6_+_46668717 | 0.04 |
ENSDART00000064853
|
oxtrl
|
oxytocin receptor like |
| chr4_+_69577362 | 0.04 |
ENSDART00000160913
|
znf1030
|
zinc finger protein 1030 |
| chr6_-_49655879 | 0.04 |
ENSDART00000191594
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr4_-_59159690 | 0.03 |
ENSDART00000164706
|
znf1149
|
zinc finger protein 1149 |
| chr23_+_37185247 | 0.03 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr18_+_13236255 | 0.03 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr4_-_43071336 | 0.02 |
ENSDART00000150773
|
znf1072
|
zinc finger protein 1072 |
| chr21_+_28535203 | 0.02 |
ENSDART00000184950
|
CR749775.2
|
|
| chr6_-_21534301 | 0.02 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr17_+_5895500 | 0.02 |
ENSDART00000019905
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr1_-_52292235 | 0.01 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr12_-_43982343 | 0.01 |
ENSDART00000161539
|
CR385054.1
|
|
| chr4_-_39942347 | 0.01 |
ENSDART00000124410
|
znf979
|
zinc finger protein 979 |
| chr4_+_39055027 | 0.01 |
ENSDART00000150397
|
znf977
|
zinc finger protein 977 |
| chr12_-_26423439 | 0.00 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.6 | 2.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 1.3 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.4 | 2.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.4 | 1.1 | GO:0009750 | response to fructose(GO:0009750) |
| 0.3 | 2.0 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 0.8 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 1.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.2 | 1.2 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 0.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 2.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 1.3 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 1.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.3 | GO:0072068 | proximal straight tubule development(GO:0072020) distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.1 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 2.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.2 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 0.4 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.2 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.0 | 0.2 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.4 | GO:0042987 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 14.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.6 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.0 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.7 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.5 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.0 | 0.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.0 | GO:0060137 | parturition(GO:0007567) maternal process involved in parturition(GO:0060137) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.3 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 1.6 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 2.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 0.9 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.2 | 1.2 | GO:0051903 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.2 | 1.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 2.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 3.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 0.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 2.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.4 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 1.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 1.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.0 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 1.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 15.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |