Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
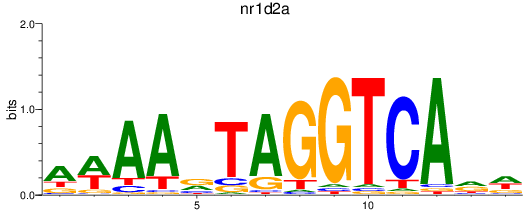
Results for nr1d2a
Z-value: 0.28
Transcription factors associated with nr1d2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1d2a
|
ENSDARG00000003820 | nuclear receptor subfamily 1, group D, member 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr1d2a | dr11_v1_chr16_+_50089417_50089417 | -0.59 | 1.0e-02 | Click! |
Activity profile of nr1d2a motif
Sorted Z-values of nr1d2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_17918810 | 0.86 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr25_-_17918536 | 0.85 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr15_+_12435975 | 0.66 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr11_-_39118882 | 0.60 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr3_-_26190804 | 0.49 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr8_+_21225064 | 0.48 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr17_-_29271359 | 0.47 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr4_-_9780931 | 0.47 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr16_-_29387215 | 0.46 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr8_+_42917515 | 0.45 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr22_+_17828267 | 0.43 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr7_+_22313533 | 0.43 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr17_+_14965570 | 0.42 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr22_-_6420239 | 0.42 |
ENSDART00000148385
|
zgc:171699
|
zgc:171699 |
| chr10_+_15454745 | 0.42 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr12_+_30789611 | 0.41 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr16_+_26612401 | 0.38 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr2_-_42558549 | 0.38 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr5_-_24029228 | 0.36 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr6_-_40768654 | 0.36 |
ENSDART00000184668
ENSDART00000146470 |
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr8_-_18899427 | 0.36 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr21_-_32781612 | 0.34 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr4_-_7875808 | 0.34 |
ENSDART00000162276
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr22_-_18546241 | 0.32 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr1_-_21714025 | 0.32 |
ENSDART00000129066
|
zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr10_+_28160265 | 0.31 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr8_-_25771474 | 0.30 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr24_-_23716097 | 0.30 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr16_-_47427016 | 0.27 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr20_+_46620374 | 0.26 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr17_-_15528597 | 0.26 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr15_-_34878388 | 0.25 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr9_-_43207768 | 0.25 |
ENSDART00000192523
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr23_+_41831224 | 0.24 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr3_+_34985895 | 0.24 |
ENSDART00000121981
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr12_-_7234915 | 0.24 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr12_+_1286642 | 0.24 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr23_-_27442544 | 0.23 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr19_+_24896409 | 0.23 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr16_+_26738404 | 0.23 |
ENSDART00000163110
|
fsbp
|
fibrinogen silencer binding protein |
| chr15_+_2559875 | 0.22 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr8_-_2602572 | 0.21 |
ENSDART00000110482
|
zdhhc12a
|
zinc finger, DHHC-type containing 12a |
| chr15_+_17100697 | 0.20 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr10_+_44692272 | 0.20 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr19_+_18797623 | 0.20 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_5666325 | 0.20 |
ENSDART00000092270
|
r3hdm4
|
R3H domain containing 4 |
| chr6_-_8244474 | 0.20 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr5_-_11573490 | 0.20 |
ENSDART00000109577
|
FO704871.1
|
|
| chr23_+_44427188 | 0.19 |
ENSDART00000160207
|
eif4e2rs1
|
eukaryotic translation initiation factor 4E family member 2 related sequence 1 |
| chr17_+_49500820 | 0.19 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr22_+_786556 | 0.19 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr22_-_15704704 | 0.19 |
ENSDART00000017838
ENSDART00000130238 |
safb
|
scaffold attachment factor B |
| chr6_-_21726758 | 0.18 |
ENSDART00000083085
|
mtmr14
|
myotubularin related protein 14 |
| chr6_-_45995401 | 0.18 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr5_-_13778726 | 0.18 |
ENSDART00000051655
|
snrnp27
|
small nuclear ribonucleoprotein 27 (U4/U6.U5) |
| chr6_+_37752781 | 0.18 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr24_+_31379318 | 0.18 |
ENSDART00000183707
|
cpne3
|
copine III |
| chr9_+_28116079 | 0.18 |
ENSDART00000101338
|
idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr21_+_26606741 | 0.17 |
ENSDART00000002767
|
esrra
|
estrogen-related receptor alpha |
| chr8_+_2530065 | 0.17 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr3_+_32698424 | 0.17 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr5_+_43470544 | 0.17 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr12_+_23912074 | 0.16 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr8_+_21146262 | 0.15 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr18_-_42313798 | 0.15 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr9_-_43213229 | 0.15 |
ENSDART00000139775
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr12_-_979789 | 0.14 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr23_-_5683147 | 0.14 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr15_+_17100412 | 0.14 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr2_-_37743834 | 0.14 |
ENSDART00000088040
ENSDART00000191057 |
myo9b
|
myosin IXb |
| chr21_-_40557281 | 0.13 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr9_+_17982737 | 0.12 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr21_-_40049642 | 0.12 |
ENSDART00000124416
|
med31
|
mediator complex subunit 31 |
| chr6_-_49159207 | 0.12 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr9_-_43213057 | 0.11 |
ENSDART00000059448
ENSDART00000133589 |
sestd1
|
SEC14 and spectrin domains 1 |
| chr18_+_6162194 | 0.11 |
ENSDART00000177153
|
otogl
|
otogelin-like |
| chr19_-_10295173 | 0.11 |
ENSDART00000136697
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr3_+_52684556 | 0.10 |
ENSDART00000139037
|
pgls
|
6-phosphogluconolactonase |
| chr15_-_31419805 | 0.10 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr13_-_9111927 | 0.10 |
ENSDART00000133815
ENSDART00000109783 ENSDART00000142540 ENSDART00000192613 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-19p15.4 si:dkey-112g5.12 |
| chr10_+_9561066 | 0.09 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr8_+_23034718 | 0.09 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr18_+_907266 | 0.09 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr23_-_36003282 | 0.09 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr2_+_35733335 | 0.08 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr7_-_28058442 | 0.07 |
ENSDART00000173842
|
si:ch211-235p24.2
|
si:ch211-235p24.2 |
| chr5_-_65021736 | 0.05 |
ENSDART00000162368
ENSDART00000161876 |
anxa1c
|
annexin A1c |
| chr9_+_35860975 | 0.05 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr10_-_25628555 | 0.05 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr18_-_39473055 | 0.05 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr22_+_36704764 | 0.05 |
ENSDART00000146662
|
trim35-33
|
tripartite motif containing 35-33 |
| chr21_+_4348600 | 0.05 |
ENSDART00000138463
|
HTRA2 (1 of many)
|
si:dkey-84o3.2 |
| chr9_+_33267211 | 0.05 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr8_+_14890821 | 0.04 |
ENSDART00000190966
|
soat1
|
sterol O-acyltransferase 1 |
| chr15_+_19681718 | 0.03 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr24_-_32408404 | 0.03 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr18_-_15559817 | 0.03 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr6_+_32415132 | 0.02 |
ENSDART00000155790
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr2_+_58841181 | 0.02 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr20_+_46191630 | 0.02 |
ENSDART00000100520
|
taar13d
|
trace amine associated receptor 13d |
| chr2_-_8648440 | 0.02 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr18_+_33570170 | 0.01 |
ENSDART00000133276
|
si:dkey-47k20.3
|
si:dkey-47k20.3 |
| chr11_-_21303946 | 0.01 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr12_-_9533956 | 0.01 |
ENSDART00000188940
|
nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr20_-_9980318 | 0.01 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr10_-_7555660 | 0.01 |
ENSDART00000163689
|
wrn
|
Werner syndrome |
| chr2_-_38337122 | 0.01 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr3_+_60761811 | 0.01 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr3_+_57997980 | 0.01 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr1_-_33353778 | 0.01 |
ENSDART00000041191
|
gyg2
|
glycogenin 2 |
| chr19_+_4990496 | 0.01 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr15_+_19682013 | 0.00 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr15_+_17030941 | 0.00 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr10_-_7555326 | 0.00 |
ENSDART00000162191
ENSDART00000186945 |
wrn
|
Werner syndrome |
| chr14_-_42997145 | 0.00 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr1d2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.3 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 0.3 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.2 | GO:1903428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.2 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.1 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.9 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.2 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.2 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |