Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
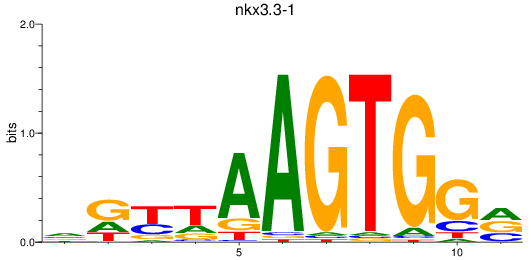
Results for nkx3.3-1
Z-value: 0.58
Transcription factors associated with nkx3.3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3.3-1
|
ENSDARG00000110589 | NK3 homeobox 3 |
Activity profile of nkx3.3-1 motif
Sorted Z-values of nkx3.3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_9467049 | 1.63 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr6_-_31739709 | 1.44 |
ENSDART00000087964
|
cachd1
|
cache domain containing 1 |
| chr24_-_33308045 | 1.34 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr9_+_8380728 | 1.29 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr3_-_60886984 | 1.20 |
ENSDART00000170974
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr20_+_23498255 | 1.18 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr19_-_43757568 | 1.12 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr6_-_1820606 | 1.08 |
ENSDART00000183228
|
FO834857.1
|
|
| chr13_+_33462232 | 1.08 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr6_-_31987940 | 1.02 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr23_+_25292147 | 0.99 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr7_-_30272871 | 0.99 |
ENSDART00000099586
|
zgc:162945
|
zgc:162945 |
| chr25_+_2361721 | 0.95 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr5_+_44846280 | 0.94 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr15_+_45595385 | 0.93 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr22_+_724639 | 0.92 |
ENSDART00000105323
|
zgc:162255
|
zgc:162255 |
| chr2_-_57900430 | 0.92 |
ENSDART00000132245
ENSDART00000140060 |
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr1_-_58887610 | 0.91 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr19_-_11031145 | 0.90 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr10_+_10387328 | 0.90 |
ENSDART00000080904
|
sardh
|
sarcosine dehydrogenase |
| chr7_+_36467796 | 0.87 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr10_+_10386435 | 0.87 |
ENSDART00000179214
ENSDART00000189799 ENSDART00000193875 |
sardh
|
sarcosine dehydrogenase |
| chr5_-_54712159 | 0.85 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr11_+_19370717 | 0.77 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr16_+_46725087 | 0.73 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
| chr4_-_10835620 | 0.73 |
ENSDART00000150739
|
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr13_-_50463938 | 0.72 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr3_+_11926030 | 0.72 |
ENSDART00000081367
|
dnaja3a
|
DnaJ (Hsp40) homolog, subfamily A, member 3A |
| chr8_+_10869183 | 0.70 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr5_-_57820873 | 0.70 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr2_-_57918314 | 0.69 |
ENSDART00000138265
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr2_-_55797318 | 0.68 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr19_+_791538 | 0.67 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr14_-_41478265 | 0.66 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr5_-_38248347 | 0.66 |
ENSDART00000084917
ENSDART00000139479 |
slc12a9
|
solute carrier family 12, member 9 |
| chr25_+_28282274 | 0.66 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr3_-_52899394 | 0.65 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr14_+_21722235 | 0.63 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr6_-_41135215 | 0.62 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr5_-_55848511 | 0.60 |
ENSDART00000183503
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr23_+_12840080 | 0.59 |
ENSDART00000081016
ENSDART00000121697 |
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr4_+_13931578 | 0.59 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr20_+_46741074 | 0.59 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr12_+_16087077 | 0.59 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr15_+_2475894 | 0.59 |
ENSDART00000035939
|
panx1a
|
pannexin 1a |
| chr19_-_29302249 | 0.57 |
ENSDART00000188751
|
srfbp1
|
serum response factor binding protein 1 |
| chr19_-_6840506 | 0.56 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr12_-_3053699 | 0.55 |
ENSDART00000139721
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr23_-_26227805 | 0.54 |
ENSDART00000158082
|
BX927204.1
|
|
| chr23_-_29812667 | 0.54 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr11_+_19271557 | 0.52 |
ENSDART00000190559
|
prickle2b
|
prickle homolog 2b |
| chr8_-_14554785 | 0.51 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_-_1604601 | 0.51 |
ENSDART00000143130
|
agps
|
alkylglycerone phosphate synthase |
| chr23_-_24542952 | 0.51 |
ENSDART00000088777
|
atp13a2
|
ATPase 13A2 |
| chr12_+_22580579 | 0.50 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr11_+_19370447 | 0.49 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr15_+_20281305 | 0.49 |
ENSDART00000155065
|
plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr16_-_26132122 | 0.48 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr24_-_25166720 | 0.48 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr19_+_32321797 | 0.47 |
ENSDART00000167664
|
atxn1a
|
ataxin 1a |
| chr21_-_21527983 | 0.47 |
ENSDART00000014254
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr16_+_38240027 | 0.47 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr4_-_13931508 | 0.47 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr22_+_32228882 | 0.46 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr23_-_18568522 | 0.46 |
ENSDART00000004655
|
sephs2
|
selenophosphate synthetase 2 |
| chr20_+_34151670 | 0.46 |
ENSDART00000152870
ENSDART00000010329 ENSDART00000145852 |
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr18_-_22094102 | 0.45 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr21_-_30026359 | 0.44 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr9_-_34882516 | 0.44 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr25_+_33033633 | 0.44 |
ENSDART00000192336
|
tln2b
|
talin 2b |
| chr7_-_40578733 | 0.44 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr10_-_38456382 | 0.44 |
ENSDART00000182129
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr15_-_20709289 | 0.43 |
ENSDART00000136767
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr16_-_22585289 | 0.43 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr1_-_30568225 | 0.43 |
ENSDART00000144297
ENSDART00000164204 |
ubac2
|
UBA domain containing 2 |
| chr17_-_51262430 | 0.43 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr11_+_31680513 | 0.43 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr1_-_49950643 | 0.43 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr4_-_73756673 | 0.42 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr5_-_26764880 | 0.42 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr8_-_25814263 | 0.41 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr19_-_34970312 | 0.39 |
ENSDART00000102896
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr23_-_21534455 | 0.39 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr5_-_26765188 | 0.39 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr16_+_25137483 | 0.39 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr23_-_21534738 | 0.39 |
ENSDART00000134587
|
rcc2
|
regulator of chromosome condensation 2 |
| chr7_-_23777445 | 0.38 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr5_-_23117078 | 0.38 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr5_-_57204352 | 0.36 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr7_+_20260172 | 0.36 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr20_-_28698172 | 0.35 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr12_-_26022663 | 0.35 |
ENSDART00000166769
|
bmpr1ab
|
bone morphogenetic protein receptor, type IAb |
| chr3_-_18384501 | 0.34 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr2_-_9527129 | 0.34 |
ENSDART00000157422
ENSDART00000004398 |
cope
|
coatomer protein complex, subunit epsilon |
| chr12_+_27243059 | 0.33 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr20_-_53078607 | 0.33 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr3_-_34753605 | 0.32 |
ENSDART00000000160
|
thraa
|
thyroid hormone receptor alpha a |
| chr11_+_23933016 | 0.32 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr6_+_58622831 | 0.31 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr13_-_35892051 | 0.31 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr22_+_2830703 | 0.31 |
ENSDART00000145463
ENSDART00000144785 |
si:dkey-20i20.8
|
si:dkey-20i20.8 |
| chr10_-_38468847 | 0.30 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr18_+_22174630 | 0.30 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr19_+_8612839 | 0.30 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr23_-_21535040 | 0.30 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr9_-_7390388 | 0.30 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr17_+_23554932 | 0.29 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr15_+_15771418 | 0.28 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr9_+_51655636 | 0.28 |
ENSDART00000169908
|
RBMS1 (1 of many)
|
RNA binding motif single stranded interacting protein 1 |
| chr3_-_31069776 | 0.28 |
ENSDART00000167462
|
elob
|
elongin B |
| chr16_+_3982590 | 0.28 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr14_+_49152341 | 0.27 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr19_-_35155722 | 0.27 |
ENSDART00000151924
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr19_+_46222428 | 0.27 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr14_+_26759332 | 0.27 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr2_-_11504778 | 0.27 |
ENSDART00000186556
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr25_-_37284370 | 0.26 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr23_-_17003533 | 0.26 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr13_+_23093743 | 0.26 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr1_-_26045560 | 0.26 |
ENSDART00000172737
ENSDART00000076120 ENSDART00000193593 |
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr25_+_25124684 | 0.25 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr20_-_33675676 | 0.25 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr5_+_57726425 | 0.25 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr19_+_40248697 | 0.25 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr23_+_25172976 | 0.25 |
ENSDART00000140789
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr22_-_16755885 | 0.25 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr17_-_16069905 | 0.24 |
ENSDART00000110383
|
map7a
|
microtubule-associated protein 7a |
| chr11_-_36446655 | 0.24 |
ENSDART00000179025
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr14_-_51014292 | 0.24 |
ENSDART00000029797
|
faf2
|
Fas associated factor family member 2 |
| chr19_+_3206263 | 0.23 |
ENSDART00000020344
|
zgc:86598
|
zgc:86598 |
| chr13_+_27040887 | 0.23 |
ENSDART00000132714
|
soul2
|
heme-binding protein soul2 |
| chr21_-_2158298 | 0.23 |
ENSDART00000182199
|
gb:ai877918
|
expressed sequence AI877918 |
| chr2_-_5199431 | 0.23 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr17_+_15388479 | 0.23 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr2_+_24868010 | 0.23 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr3_+_14581643 | 0.23 |
ENSDART00000182840
|
znf653
|
zinc finger protein 653 |
| chr6_-_52235118 | 0.22 |
ENSDART00000191243
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr11_-_5953636 | 0.21 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr10_-_29831944 | 0.21 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr3_-_41795917 | 0.21 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr25_+_10547228 | 0.21 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr22_+_28803739 | 0.21 |
ENSDART00000129476
ENSDART00000189726 |
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr2_+_30463825 | 0.21 |
ENSDART00000092356
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr12_+_18458502 | 0.20 |
ENSDART00000108745
|
rnf151
|
ring finger protein 151 |
| chr8_-_51507144 | 0.20 |
ENSDART00000024882
ENSDART00000135166 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr7_-_5487593 | 0.20 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr4_-_2059233 | 0.20 |
ENSDART00000188177
ENSDART00000129521 ENSDART00000082289 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr21_-_2162850 | 0.20 |
ENSDART00000159731
|
gb:ai877918
|
expressed sequence AI877918 |
| chr8_+_24281512 | 0.19 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr21_+_34814444 | 0.18 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr10_-_33343244 | 0.18 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr25_+_10416583 | 0.18 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr14_-_31814149 | 0.18 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr8_+_49766338 | 0.18 |
ENSDART00000060657
|
rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chr11_-_2221159 | 0.18 |
ENSDART00000189292
|
smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr5_+_13870340 | 0.18 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr2_+_51028269 | 0.17 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr13_+_18371208 | 0.17 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr12_+_17754859 | 0.17 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr10_+_35358675 | 0.17 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr8_+_23788981 | 0.17 |
ENSDART00000144229
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr22_-_31020690 | 0.16 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr7_+_17063761 | 0.16 |
ENSDART00000182880
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr6_+_41808673 | 0.16 |
ENSDART00000038163
|
rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr7_+_50796422 | 0.16 |
ENSDART00000123868
|
dthd1
|
death domain containing 1 |
| chr5_+_37890521 | 0.15 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr10_-_39052264 | 0.15 |
ENSDART00000144036
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr10_+_37268854 | 0.15 |
ENSDART00000131897
|
nf1b
|
neurofibromin 1b |
| chr6_-_59381391 | 0.14 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr17_-_25831569 | 0.14 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr8_-_43689324 | 0.13 |
ENSDART00000159904
|
ep400
|
E1A binding protein p400 |
| chr4_-_20521441 | 0.12 |
ENSDART00000066895
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr1_+_35862550 | 0.12 |
ENSDART00000132118
|
si:ch211-194g2.4
|
si:ch211-194g2.4 |
| chr12_+_2381213 | 0.12 |
ENSDART00000188007
|
LO018238.1
|
|
| chr10_+_40684758 | 0.12 |
ENSDART00000133648
|
taar19h
|
trace amine associated receptor 19h |
| chr4_-_5077158 | 0.12 |
ENSDART00000155915
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr23_+_7518294 | 0.11 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr21_+_1380099 | 0.11 |
ENSDART00000184516
|
TCF4
|
transcription factor 4 |
| chr20_+_44311448 | 0.10 |
ENSDART00000114660
|
opn8b
|
opsin 8, group member b |
| chr22_+_19188809 | 0.10 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr16_-_9675982 | 0.10 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr3_-_5393909 | 0.10 |
ENSDART00000031885
|
si:ch73-106l15.2
|
si:ch73-106l15.2 |
| chr18_+_33675664 | 0.10 |
ENSDART00000140043
|
si:dkey-47k20.8
|
si:dkey-47k20.8 |
| chr19_+_43256986 | 0.09 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr4_+_19534833 | 0.09 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr1_-_11519934 | 0.09 |
ENSDART00000162060
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr21_+_39948300 | 0.09 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr14_+_26719691 | 0.09 |
ENSDART00000078522
ENSDART00000172927 |
eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr16_-_38609146 | 0.09 |
ENSDART00000144651
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr16_+_49647402 | 0.09 |
ENSDART00000015694
ENSDART00000132547 |
rab5ab
|
RAB5A, member RAS oncogene family, b |
| chr4_-_75982054 | 0.08 |
ENSDART00000168369
|
si:ch211-232d10.1
|
si:ch211-232d10.1 |
| chr20_-_46053720 | 0.08 |
ENSDART00000136254
|
taar12h
|
trace amine associated receptor 12h |
| chr4_-_62503772 | 0.08 |
ENSDART00000108891
|
si:dkey-165b20.1
|
si:dkey-165b20.1 |
| chr6_+_13606410 | 0.08 |
ENSDART00000104716
|
asic4b
|
acid-sensing (proton-gated) ion channel family member 4b |
| chr6_+_47424266 | 0.08 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr5_-_57655092 | 0.08 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr2_+_30281444 | 0.08 |
ENSDART00000172756
|
si:dkey-82k12.13
|
si:dkey-82k12.13 |
| chr8_+_22289320 | 0.08 |
ENSDART00000075126
|
b3gnt7l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like |
| chr25_+_20091021 | 0.08 |
ENSDART00000187545
ENSDART00000053265 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr4_-_71708567 | 0.08 |
ENSDART00000182645
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3.3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.2 | 0.7 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.2 | 0.9 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 0.6 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 0.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.3 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.9 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.4 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 1.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.7 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.2 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.1 | 0.2 | GO:1990120 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.3 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 1.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.0 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.4 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.5 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.6 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.2 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.4 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.0 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0060959 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.5 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 1.4 | GO:1903706 | regulation of hemopoiesis(GO:1903706) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.7 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 0.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.2 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.0 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |