Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
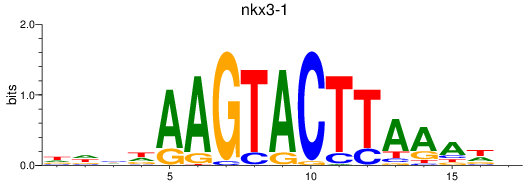
Results for nkx3-1
Z-value: 1.03
Transcription factors associated with nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-1
|
ENSDARG00000078280 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3-1 | dr11_v1_chr8_-_50259448_50259448 | -0.90 | 3.1e-07 | Click! |
Activity profile of nkx3-1 motif
Sorted Z-values of nkx3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_8396755 | 5.57 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr22_+_25236657 | 5.41 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr5_+_16117871 | 4.70 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr9_+_44994214 | 4.52 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr22_+_25249193 | 4.16 |
ENSDART00000171851
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr10_-_34915886 | 3.37 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr10_-_34916208 | 3.08 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr7_+_17947217 | 2.74 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr15_+_29025090 | 1.97 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr3_-_31875138 | 1.96 |
ENSDART00000155298
|
limd2
|
LIM domain containing 2 |
| chr22_-_10541712 | 1.96 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr17_-_43031763 | 1.95 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr5_-_10082244 | 1.94 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr16_+_54641230 | 1.92 |
ENSDART00000157641
ENSDART00000159540 |
fbxo43
|
F-box protein 43 |
| chr9_-_8296723 | 1.86 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr9_-_12888082 | 1.84 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr16_-_42175617 | 1.77 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr1_-_55248496 | 1.76 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr7_-_26532089 | 1.76 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr12_+_14079097 | 1.70 |
ENSDART00000078033
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr15_-_4415917 | 1.70 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr7_+_10911396 | 1.63 |
ENSDART00000167273
ENSDART00000081323 ENSDART00000170655 |
abhd17c
|
abhydrolase domain containing 17C |
| chr6_+_10338554 | 1.62 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr24_-_31904924 | 1.60 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr4_+_3312852 | 1.59 |
ENSDART00000084691
|
fgd4b
|
FYVE, RhoGEF and PH domain containing 4b |
| chr5_+_36666715 | 1.57 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr11_-_23697217 | 1.52 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr19_-_47587719 | 1.47 |
ENSDART00000111108
|
CABZ01071972.1
|
|
| chr19_-_31707892 | 1.45 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr20_-_13625588 | 1.43 |
ENSDART00000078893
|
sytl3
|
synaptotagmin-like 3 |
| chr10_+_585719 | 1.40 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr10_+_3153973 | 1.35 |
ENSDART00000183223
|
hic2
|
hypermethylated in cancer 2 |
| chr6_-_49537646 | 1.34 |
ENSDART00000180438
|
FO704848.1
|
|
| chr10_-_3427589 | 1.32 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr3_-_26806032 | 1.30 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_+_22632126 | 1.30 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr13_-_36680531 | 1.29 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr8_-_16675464 | 1.29 |
ENSDART00000191982
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr7_-_58098814 | 1.25 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr21_+_18907102 | 1.24 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr7_-_26518086 | 1.22 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr12_-_10476448 | 1.22 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr17_-_18898115 | 1.21 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr14_+_14806851 | 1.19 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr13_-_32726178 | 1.19 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr3_-_26805455 | 1.18 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr5_+_27137473 | 1.15 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr1_-_354115 | 1.13 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr24_-_32522587 | 1.13 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr24_-_26518972 | 1.12 |
ENSDART00000097792
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr8_-_38317914 | 1.10 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr16_-_41535690 | 1.09 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr23_+_27782071 | 1.09 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr19_+_32401278 | 1.08 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr16_+_16253710 | 1.07 |
ENSDART00000137672
|
setd2
|
SET domain containing 2 |
| chr15_-_28805493 | 1.07 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr21_-_11286483 | 1.03 |
ENSDART00000074845
|
rtkn2b
|
rhotekin 2b |
| chr17_-_43623356 | 1.02 |
ENSDART00000075596
ENSDART00000136431 |
rtkn2a
|
rhotekin 2a |
| chr7_-_41013575 | 1.02 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr17_-_24703778 | 1.02 |
ENSDART00000156061
|
si:ch211-15d5.12
|
si:ch211-15d5.12 |
| chr2_+_11031360 | 1.01 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr9_+_48123224 | 1.00 |
ENSDART00000141610
|
klhl23
|
kelch-like family member 23 |
| chr12_+_23850661 | 0.99 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr24_+_24086491 | 0.98 |
ENSDART00000145092
|
lipib
|
lipase, member Ib |
| chr2_+_15100742 | 0.97 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr9_-_51563575 | 0.97 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr9_-_46072805 | 0.97 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr9_+_48123002 | 0.97 |
ENSDART00000099794
ENSDART00000169733 |
klhl23
|
kelch-like family member 23 |
| chr21_-_2042037 | 0.94 |
ENSDART00000171131
ENSDART00000160144 |
add1
|
adducin 1 (alpha) |
| chr21_-_35325466 | 0.94 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr17_+_22577472 | 0.93 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr24_+_26328787 | 0.92 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr2_-_29923630 | 0.89 |
ENSDART00000158844
ENSDART00000031130 |
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr7_-_64770456 | 0.89 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr24_+_16140423 | 0.89 |
ENSDART00000105955
|
TIMM21
|
si:dkey-118j18.1 |
| chr9_-_52598343 | 0.88 |
ENSDART00000167922
|
xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr25_+_34862225 | 0.85 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr9_-_14273652 | 0.85 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr7_-_32875552 | 0.84 |
ENSDART00000132504
|
ano5b
|
anoctamin 5b |
| chr16_+_33142734 | 0.84 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr4_-_77252368 | 0.83 |
ENSDART00000111941
|
zgc:174310
|
zgc:174310 |
| chr3_+_36222799 | 0.83 |
ENSDART00000113105
|
dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr12_+_33361948 | 0.82 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr5_+_18012154 | 0.81 |
ENSDART00000139431
ENSDART00000048859 |
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr10_-_8957480 | 0.81 |
ENSDART00000150049
|
mocs2
|
molybdenum cofactor synthesis 2 |
| chr14_+_48903640 | 0.81 |
ENSDART00000165323
|
zgc:154054
|
zgc:154054 |
| chr25_+_3788074 | 0.80 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr3_-_15668433 | 0.79 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| chr20_-_25902141 | 0.78 |
ENSDART00000142611
ENSDART00000024821 |
elmsan1a
|
ELM2 and Myb/SANT-like domain containing 1a |
| chr7_-_58178980 | 0.78 |
ENSDART00000073635
|
nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr9_+_28140089 | 0.78 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr2_-_10877228 | 0.77 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr20_+_46620374 | 0.77 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr11_+_30636351 | 0.76 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr17_+_10578823 | 0.76 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr4_-_4570475 | 0.75 |
ENSDART00000184955
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr1_-_51261420 | 0.74 |
ENSDART00000168685
|
kif16ba
|
kinesin family member 16Ba |
| chr3_-_18189283 | 0.73 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr22_+_2769236 | 0.71 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr19_-_10330778 | 0.70 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr6_-_8465656 | 0.70 |
ENSDART00000178887
|
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr25_-_24240797 | 0.69 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr14_+_14806692 | 0.68 |
ENSDART00000193050
|
fhdc2
|
FH2 domain containing 2 |
| chr6_+_21992820 | 0.68 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr7_-_30280934 | 0.68 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr7_-_34192834 | 0.68 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr21_-_4250682 | 0.68 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr6_-_19351495 | 0.68 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr2_+_45696743 | 0.68 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr25_-_34973211 | 0.68 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr3_-_36420397 | 0.67 |
ENSDART00000167896
|
cog1
|
component of oligomeric golgi complex 1 |
| chr19_-_3821678 | 0.67 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr7_+_30725473 | 0.67 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr12_-_25150239 | 0.65 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr13_+_7242916 | 0.65 |
ENSDART00000184238
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr3_-_36419641 | 0.65 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr11_+_2649664 | 0.63 |
ENSDART00000166357
|
si:ch211-160o17.4
|
si:ch211-160o17.4 |
| chr23_-_4019928 | 0.62 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr23_-_4019699 | 0.62 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_+_6538733 | 0.61 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr7_+_37742299 | 0.61 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr5_-_23795688 | 0.61 |
ENSDART00000099084
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr5_+_31965845 | 0.61 |
ENSDART00000112968
|
myo1hb
|
myosin IHb |
| chr6_+_9981300 | 0.61 |
ENSDART00000151482
|
abi2b
|
abl-interactor 2b |
| chr11_+_2649891 | 0.59 |
ENSDART00000093052
|
si:ch211-160o17.4
|
si:ch211-160o17.4 |
| chr14_-_33978117 | 0.59 |
ENSDART00000128515
|
foxa
|
forkhead box A sequence |
| chr1_-_49950643 | 0.59 |
ENSDART00000138301
|
sgms2
|
sphingomyelin synthase 2 |
| chr25_+_31122806 | 0.58 |
ENSDART00000067039
|
rassf8a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8a |
| chr25_+_34749187 | 0.58 |
ENSDART00000141473
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_+_50477779 | 0.58 |
ENSDART00000122716
|
CABZ01067973.1
|
|
| chr16_-_42238120 | 0.58 |
ENSDART00000084730
|
zgc:162160
|
zgc:162160 |
| chr9_-_50001606 | 0.58 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr19_+_7424347 | 0.57 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr5_-_25576462 | 0.57 |
ENSDART00000165147
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr15_+_41836104 | 0.57 |
ENSDART00000137434
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr6_+_21993613 | 0.54 |
ENSDART00000160750
ENSDART00000083070 |
thumpd3
|
THUMP domain containing 3 |
| chr3_+_25849560 | 0.54 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr5_-_57204352 | 0.54 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr2_+_10642047 | 0.53 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr7_-_20464468 | 0.53 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr12_+_32729470 | 0.52 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr20_-_18535502 | 0.52 |
ENSDART00000049437
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr14_+_29581710 | 0.52 |
ENSDART00000188820
ENSDART00000193874 |
TENM2
|
si:dkey-34l15.2 |
| chr17_+_13664442 | 0.51 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr18_+_6558338 | 0.50 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr3_-_13599482 | 0.49 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr11_+_25157374 | 0.48 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr5_+_30596477 | 0.48 |
ENSDART00000124487
|
hinfp
|
histone H4 transcription factor |
| chr4_-_16706776 | 0.48 |
ENSDART00000079461
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr9_-_32804953 | 0.47 |
ENSDART00000134763
|
zgc:112056
|
zgc:112056 |
| chr19_-_12315693 | 0.47 |
ENSDART00000151158
|
ncaldb
|
neurocalcin delta b |
| chr4_+_76466751 | 0.47 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr25_+_22587306 | 0.47 |
ENSDART00000067479
|
stra6
|
stimulated by retinoic acid 6 |
| chr9_+_35016201 | 0.47 |
ENSDART00000182404
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr20_-_38525467 | 0.45 |
ENSDART00000061417
|
si:ch211-245h14.1
|
si:ch211-245h14.1 |
| chr21_+_19547806 | 0.45 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr8_+_46939391 | 0.43 |
ENSDART00000146631
|
espn
|
espin |
| chr21_-_14815952 | 0.42 |
ENSDART00000134278
ENSDART00000067004 |
phpt1
|
phosphohistidine phosphatase 1 |
| chr14_-_48786708 | 0.42 |
ENSDART00000169730
|
si:ch211-199b20.3
|
si:ch211-199b20.3 |
| chr15_+_17848590 | 0.42 |
ENSDART00000168940
|
zgc:113279
|
zgc:113279 |
| chr14_-_31814149 | 0.42 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_7673165 | 0.41 |
ENSDART00000028171
|
lta4h
|
leukotriene A4 hydrolase |
| chr5_+_30596632 | 0.41 |
ENSDART00000051414
|
hinfp
|
histone H4 transcription factor |
| chr3_+_32389904 | 0.41 |
ENSDART00000154788
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr18_+_35861930 | 0.40 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr2_-_27575803 | 0.40 |
ENSDART00000014568
|
urod
|
uroporphyrinogen decarboxylase |
| chr19_+_20793237 | 0.39 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr11_-_7156620 | 0.39 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr10_+_44373349 | 0.38 |
ENSDART00000172191
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr5_+_5406503 | 0.38 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr1_+_44056129 | 0.38 |
ENSDART00000161114
|
CU915778.1
|
|
| chr15_+_25452092 | 0.37 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr21_-_25623342 | 0.37 |
ENSDART00000101201
|
tmem223
|
transmembrane protein 223 |
| chr25_+_34748860 | 0.37 |
ENSDART00000087364
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr5_-_24882357 | 0.37 |
ENSDART00000127773
ENSDART00000003998 ENSDART00000143851 |
ewsr1b
|
EWS RNA-binding protein 1b |
| chr14_+_50937757 | 0.37 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr8_-_22326073 | 0.36 |
ENSDART00000084965
|
cep104
|
centrosomal protein 104 |
| chr20_-_33507458 | 0.35 |
ENSDART00000140287
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr14_+_35013656 | 0.35 |
ENSDART00000164974
|
ebf3a
|
early B cell factor 3a |
| chr1_+_32054159 | 0.33 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr5_-_55752169 | 0.33 |
ENSDART00000097424
|
kcmf1
|
potassium channel modulatory factor 1 |
| chr7_+_41322407 | 0.32 |
ENSDART00000114076
ENSDART00000139093 |
dph2
|
DPH2 homolog (S. cerevisiae) |
| chr12_-_4424424 | 0.32 |
ENSDART00000152314
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr22_+_28818291 | 0.31 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr4_+_76671012 | 0.31 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr20_+_36623807 | 0.31 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr9_-_52386733 | 0.29 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr16_+_43368572 | 0.28 |
ENSDART00000032778
ENSDART00000193897 |
rnf144b
|
ring finger protein 144B |
| chr24_+_26017094 | 0.27 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr19_+_33139164 | 0.27 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr1_-_59169815 | 0.27 |
ENSDART00000100163
|
wu:fk65c09
|
wu:fk65c09 |
| chr16_-_47427016 | 0.27 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr8_-_22826996 | 0.27 |
ENSDART00000146563
|
magixa
|
MAGI family member, X-linked a |
| chr12_-_33579873 | 0.26 |
ENSDART00000184661
|
tdrkh
|
tudor and KH domain containing |
| chr3_-_27915270 | 0.26 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr18_-_12416019 | 0.26 |
ENSDART00000144799
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr2_-_11101329 | 0.26 |
ENSDART00000164416
|
lrrc53
|
leucine rich repeat containing 53 |
| chr4_+_74928675 | 0.25 |
ENSDART00000192755
|
nup50
|
nucleoporin 50 |
| chr24_+_30521619 | 0.25 |
ENSDART00000162903
|
dpyda.3
|
dihydropyrimidine dehydrogenase a, tandem duplicate 3 |
| chr1_-_53273500 | 0.24 |
ENSDART00000150352
|
znf330
|
zinc finger protein 330 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.5 | 1.5 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.4 | 1.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 1.0 | GO:2000638 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 2.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.3 | 0.9 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.3 | 0.8 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 1.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.0 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 1.9 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.2 | 0.7 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 0.8 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.2 | 0.9 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.6 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 1.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 1.0 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.4 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 1.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.1 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.6 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.8 | GO:0002360 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 2 cell differentiation(GO:0045064) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 4.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.5 | GO:0035461 | vitamin transmembrane transport(GO:0035461) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 3.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.8 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 2.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.8 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 1.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.4 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.9 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.1 | 2.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 1.0 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.3 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 1.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0019860 | uracil catabolic process(GO:0006212) beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) uracil metabolic process(GO:0019860) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 1.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.1 | GO:0021985 | parturition(GO:0007567) neurohypophysis development(GO:0021985) multi-multicellular organism process(GO:0044706) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.4 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.4 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.1 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.7 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.0 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) positive regulation of lipid transport(GO:0032370) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.1 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.3 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.4 | 2.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 1.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.8 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 2.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 2.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 3.9 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.2 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.9 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.4 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.4 | 2.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.7 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.8 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 1.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.4 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.5 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 1.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 4.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 17.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 3.2 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 1.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.1 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 6.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 1.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.3 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |