Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
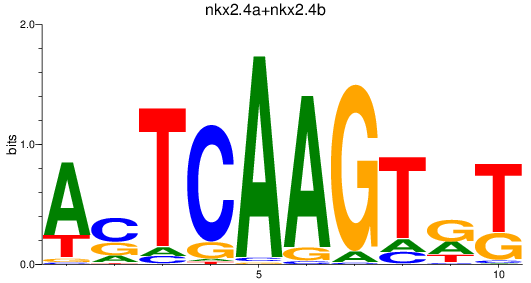
Results for nkx2.4a+nkx2.4b
Z-value: 0.32
Transcription factors associated with nkx2.4a+nkx2.4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.4a
|
ENSDARG00000075107 | NK2 homeobox 4a |
|
nkx2.4b
|
ENSDARG00000104107 | NK2 homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.4b | dr11_v1_chr20_+_48803248_48803248 | -0.64 | 3.9e-03 | Click! |
| nkx2.4a | dr11_v1_chr17_-_42097267_42097267 | -0.03 | 9.2e-01 | Click! |
Activity profile of nkx2.4a+nkx2.4b motif
Sorted Z-values of nkx2.4a+nkx2.4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_23294753 | 0.89 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr1_+_31657842 | 0.78 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr1_+_31658011 | 0.74 |
ENSDART00000192203
|
poll
|
polymerase (DNA directed), lambda |
| chr15_-_28569786 | 0.68 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr8_-_39884359 | 0.67 |
ENSDART00000131372
|
mlec
|
malectin |
| chr16_+_25259313 | 0.60 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr8_-_22558773 | 0.58 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr1_-_38171648 | 0.53 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr22_-_18241390 | 0.52 |
ENSDART00000083879
|
tmem161a
|
transmembrane protein 161A |
| chr22_-_18240968 | 0.50 |
ENSDART00000027605
|
tmem161a
|
transmembrane protein 161A |
| chr23_+_27778670 | 0.49 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr7_+_13830052 | 0.48 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr9_+_2762270 | 0.47 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr7_+_39416336 | 0.47 |
ENSDART00000171783
|
CT030188.1
|
|
| chr24_+_19210001 | 0.46 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr8_-_39859688 | 0.45 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr15_-_28904371 | 0.45 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_38419575 | 0.44 |
ENSDART00000087639
|
smap2
|
small ArfGAP2 |
| chr24_-_205275 | 0.44 |
ENSDART00000108762
|
vopp1
|
VOPP1, WBP1/VOPP1 family member |
| chr9_+_22634073 | 0.42 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr19_-_25113660 | 0.42 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr24_-_5691956 | 0.41 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr10_-_10018120 | 0.41 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr20_+_23742574 | 0.40 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| chr10_+_5268054 | 0.39 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr3_-_26805455 | 0.39 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr22_-_21897203 | 0.38 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr9_+_4378153 | 0.36 |
ENSDART00000191264
ENSDART00000182384 |
kalrna
|
kalirin RhoGEF kinase a |
| chr15_+_34963316 | 0.36 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr1_+_22654875 | 0.36 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr2_+_47754419 | 0.34 |
ENSDART00000188221
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr14_+_16083818 | 0.34 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr7_+_29163762 | 0.33 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr2_+_36608387 | 0.32 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr5_-_54554583 | 0.31 |
ENSDART00000158865
ENSDART00000158069 |
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr1_-_28950366 | 0.31 |
ENSDART00000110270
|
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr14_+_23709134 | 0.30 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr13_-_4018888 | 0.30 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr7_+_7630409 | 0.30 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr18_+_50669456 | 0.29 |
ENSDART00000163005
|
si:dkey-151j17.4
|
si:dkey-151j17.4 |
| chr1_+_45671687 | 0.28 |
ENSDART00000146101
|
mcoln1a
|
mucolipin 1a |
| chr5_-_39171302 | 0.28 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr14_+_23709543 | 0.28 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr15_-_14193926 | 0.28 |
ENSDART00000162707
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr23_-_33709964 | 0.28 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr7_+_42206847 | 0.28 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr15_-_14194208 | 0.28 |
ENSDART00000188237
ENSDART00000183155 ENSDART00000165520 |
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr10_-_15048781 | 0.28 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr15_-_23485752 | 0.27 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr12_+_27243059 | 0.27 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr2_-_38284648 | 0.27 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr12_-_22238004 | 0.27 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr23_+_4348265 | 0.26 |
ENSDART00000136287
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr19_+_32401278 | 0.26 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr23_+_4348479 | 0.26 |
ENSDART00000182425
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr7_+_39664055 | 0.25 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr13_-_48388726 | 0.25 |
ENSDART00000169473
|
fbxo11a
|
F-box protein 11a |
| chr23_+_39346774 | 0.22 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr13_-_51846224 | 0.22 |
ENSDART00000184663
|
LT631684.2
|
|
| chr19_-_17303500 | 0.22 |
ENSDART00000162355
|
sf3a3
|
splicing factor 3a, subunit 3 |
| chr8_-_23776399 | 0.21 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr19_+_40250682 | 0.21 |
ENSDART00000102888
|
cdk6
|
cyclin-dependent kinase 6 |
| chr25_-_14424406 | 0.21 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr5_-_28915130 | 0.20 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr2_-_11648731 | 0.20 |
ENSDART00000080925
|
impad1
|
inositol monophosphatase domain containing 1 |
| chr17_-_8673567 | 0.20 |
ENSDART00000192714
ENSDART00000012546 |
ctbp2a
|
C-terminal binding protein 2a |
| chr3_-_39305291 | 0.19 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr17_-_8673278 | 0.19 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr2_+_16652238 | 0.19 |
ENSDART00000091351
|
gk5
|
glycerol kinase 5 (putative) |
| chr23_-_35347714 | 0.19 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr4_+_23127104 | 0.19 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr17_-_22552678 | 0.19 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr7_+_26049818 | 0.19 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr1_+_29862074 | 0.18 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr2_+_23731194 | 0.18 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr6_+_18367388 | 0.18 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr25_-_1079417 | 0.17 |
ENSDART00000163134
|
FO907089.1
|
|
| chr22_+_8753092 | 0.17 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr19_+_23982466 | 0.16 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr23_+_39346930 | 0.16 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr20_-_43079478 | 0.16 |
ENSDART00000060982
|
snx9b
|
sorting nexin 9b |
| chr3_-_29891456 | 0.16 |
ENSDART00000151677
ENSDART00000014021 |
slc25a39
|
solute carrier family 25, member 39 |
| chr5_+_36896933 | 0.16 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr14_-_30876708 | 0.16 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr4_-_22335247 | 0.14 |
ENSDART00000192781
|
golgb1
|
golgin B1 |
| chr3_+_32571929 | 0.14 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr8_-_51293265 | 0.14 |
ENSDART00000127875
ENSDART00000181145 |
bmp1a
|
bone morphogenetic protein 1a |
| chr24_-_28259127 | 0.14 |
ENSDART00000149589
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr8_+_23485079 | 0.14 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr12_+_32199651 | 0.14 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr20_-_27190393 | 0.14 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr7_+_13639473 | 0.13 |
ENSDART00000157870
ENSDART00000032899 |
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr14_-_30876299 | 0.13 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr6_+_49028874 | 0.13 |
ENSDART00000175254
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr8_-_30316436 | 0.13 |
ENSDART00000111358
ENSDART00000167065 |
zgc:162939
|
zgc:162939 |
| chr8_-_22698651 | 0.13 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr4_+_23126558 | 0.12 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr3_-_11008532 | 0.12 |
ENSDART00000165086
|
CR382337.3
|
|
| chr10_+_9346383 | 0.12 |
ENSDART00000064975
ENSDART00000141869 |
rbm18
|
RNA binding motif protein 18 |
| chr10_+_31244619 | 0.12 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr17_+_8184649 | 0.12 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr2_-_6292510 | 0.12 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr5_+_63767376 | 0.12 |
ENSDART00000138898
|
rgs3b
|
regulator of G protein signaling 3b |
| chr13_-_13294847 | 0.11 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr5_+_56119975 | 0.10 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr24_+_33392698 | 0.10 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr10_-_9086716 | 0.09 |
ENSDART00000140095
|
arl15b
|
ADP-ribosylation factor-like 15b |
| chr5_-_33825465 | 0.09 |
ENSDART00000110645
|
dab2ipb
|
DAB2 interacting protein b |
| chr24_+_3307857 | 0.09 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr16_-_49505275 | 0.09 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr10_+_573667 | 0.09 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr19_+_26424734 | 0.08 |
ENSDART00000187402
|
npr1a
|
natriuretic peptide receptor 1a |
| chr2_-_32668668 | 0.08 |
ENSDART00000190450
|
puf60a
|
poly-U binding splicing factor a |
| chr5_+_36611128 | 0.07 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr11_+_22109887 | 0.07 |
ENSDART00000122136
|
MDFI
|
si:dkey-91m3.1 |
| chr14_+_23158021 | 0.07 |
ENSDART00000084664
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr22_-_7457247 | 0.06 |
ENSDART00000106081
|
BX511034.2
|
|
| chr7_-_26049282 | 0.06 |
ENSDART00000136389
ENSDART00000101124 |
rnaseka
|
ribonuclease, RNase K a |
| chr19_+_7938121 | 0.06 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr14_-_32486757 | 0.05 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr21_+_17110598 | 0.05 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr8_+_53120278 | 0.05 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr17_-_28097760 | 0.05 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr4_+_1146346 | 0.05 |
ENSDART00000166467
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr12_+_19208338 | 0.05 |
ENSDART00000110415
|
sh3bp1
|
SH3-domain binding protein 1 |
| chr18_-_7481036 | 0.05 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr13_-_42530656 | 0.05 |
ENSDART00000084327
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr24_-_13272842 | 0.04 |
ENSDART00000146790
ENSDART00000066703 |
rdh10a
|
retinol dehydrogenase 10a |
| chr23_+_34321237 | 0.04 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr7_-_18601206 | 0.04 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr9_+_25832329 | 0.04 |
ENSDART00000130059
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr23_+_42254960 | 0.04 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr13_-_2112008 | 0.04 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr3_+_33340939 | 0.04 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr1_-_59232267 | 0.03 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr1_-_34612613 | 0.03 |
ENSDART00000004361
|
klf5a
|
Kruppel-like factor 5a |
| chr12_-_11768889 | 0.03 |
ENSDART00000057866
|
atoh1c
|
atonal bHLH transcription factor 1c |
| chr17_+_27456804 | 0.03 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr23_-_18982499 | 0.03 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr2_-_24407732 | 0.03 |
ENSDART00000180612
ENSDART00000153688 ENSDART00000179913 |
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr16_+_23924040 | 0.02 |
ENSDART00000161124
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr16_-_34258931 | 0.02 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr15_+_17441734 | 0.02 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr8_-_49345388 | 0.02 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr1_-_14234076 | 0.02 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr14_-_9982603 | 0.02 |
ENSDART00000054687
|
il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr15_-_8517555 | 0.02 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr21_+_30084823 | 0.02 |
ENSDART00000154573
|
prob1
|
proline-rich basic protein 1 |
| chr17_-_32698503 | 0.01 |
ENSDART00000157175
ENSDART00000077439 |
kcnk2a
|
potassium channel, subfamily K, member 2a |
| chr8_-_47152001 | 0.01 |
ENSDART00000163922
ENSDART00000110512 ENSDART00000024320 |
ybx1
|
Y box binding protein 1 |
| chr4_+_54618332 | 0.01 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr15_-_26552652 | 0.01 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr6_-_46782176 | 0.01 |
ENSDART00000185559
|
igfn1.4
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 4 |
| chr21_-_41147818 | 0.01 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr20_+_35484070 | 0.01 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr8_-_29658171 | 0.01 |
ENSDART00000147755
|
mpeg1.3
|
macrophage expressed 1, tandem duplicate 3 |
| chr4_-_60312069 | 0.00 |
ENSDART00000167207
|
si:dkey-248e17.4
|
si:dkey-248e17.4 |
| chr3_+_4047789 | 0.00 |
ENSDART00000148498
|
si:dkeyp-52c3.5
|
si:dkeyp-52c3.5 |
| chr25_+_19485198 | 0.00 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr6_-_42073367 | 0.00 |
ENSDART00000154304
|
grm2a
|
glutamate receptor, metabotropic 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.4a+nkx2.4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.2 | 0.6 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.4 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.9 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.5 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.4 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 0.2 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.3 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.4 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.2 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0010459 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.6 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 0.5 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.7 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |