Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
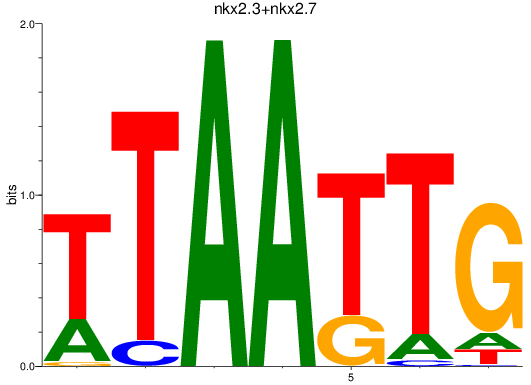
Results for nkx2.3+nkx2.7
Z-value: 1.08
Transcription factors associated with nkx2.3+nkx2.7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.7
|
ENSDARG00000021232 | NK2 transcription factor related 7 |
|
nkx2.3
|
ENSDARG00000039095 | NK2 homeobox 3 |
|
nkx2.3
|
ENSDARG00000113490 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.3 | dr11_v1_chr13_-_40411908_40411917 | -0.88 | 1.4e-06 | Click! |
| nkx2.7 | dr11_v1_chr8_-_50287949_50287949 | -0.51 | 2.9e-02 | Click! |
Activity profile of nkx2.3+nkx2.7 motif
Sorted Z-values of nkx2.3+nkx2.7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_21001264 | 3.78 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr23_+_28322986 | 3.48 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr19_-_868187 | 3.36 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr11_-_6452444 | 2.90 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr8_+_45334255 | 2.86 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr10_-_34002185 | 2.47 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr19_+_42886413 | 2.45 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr5_-_29512538 | 2.44 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr23_-_33558161 | 2.16 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr7_-_51727760 | 2.15 |
ENSDART00000174180
|
hdac8
|
histone deacetylase 8 |
| chr1_-_45213565 | 2.14 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr15_-_35112937 | 2.11 |
ENSDART00000154565
ENSDART00000099642 |
zgc:77118
|
zgc:77118 |
| chr7_+_27603211 | 2.10 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr22_-_22147375 | 2.09 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr14_-_14687004 | 2.06 |
ENSDART00000169970
|
gcna
|
germ cell nuclear acidic peptidase |
| chr5_-_23596339 | 1.88 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr23_+_9508538 | 1.87 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr4_-_4259079 | 1.86 |
ENSDART00000135352
ENSDART00000026559 |
cd9b
|
CD9 molecule b |
| chr1_-_6028876 | 1.81 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr17_+_43867889 | 1.81 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr11_+_11303458 | 1.80 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr16_-_7793457 | 1.78 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr24_+_12835935 | 1.76 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr16_-_34195002 | 1.69 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr12_+_22407852 | 1.67 |
ENSDART00000178840
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr5_+_25304499 | 1.66 |
ENSDART00000163425
|
carnmt1
|
carnosine N-methyltransferase 1 |
| chr8_-_19467011 | 1.62 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr7_-_26270014 | 1.61 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr25_-_29074064 | 1.60 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr12_-_26538823 | 1.60 |
ENSDART00000143213
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr22_-_14272699 | 1.55 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr17_+_19630272 | 1.55 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr12_-_10508952 | 1.53 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr18_-_35407695 | 1.53 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr6_+_4528631 | 1.50 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr7_-_39360325 | 1.49 |
ENSDART00000098033
ENSDART00000173695 ENSDART00000173466 ENSDART00000173734 |
ambra1a
|
autophagy/beclin-1 regulator 1a |
| chr23_-_36303216 | 1.49 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr17_+_12075805 | 1.47 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr18_-_35407289 | 1.47 |
ENSDART00000012018
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr18_-_35407530 | 1.46 |
ENSDART00000137663
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr8_-_20230559 | 1.46 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr9_-_22057658 | 1.43 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr14_+_29609245 | 1.43 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr5_-_65662996 | 1.43 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr19_+_20201254 | 1.42 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr13_+_25397098 | 1.42 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr5_+_36896933 | 1.41 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr7_+_24023653 | 1.40 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr21_+_34088110 | 1.40 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr9_+_32178050 | 1.40 |
ENSDART00000169526
|
coq10b
|
coenzyme Q10B |
| chr13_+_25396896 | 1.39 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr7_-_26497947 | 1.38 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr8_-_20230802 | 1.38 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr4_-_837768 | 1.38 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr7_+_39418869 | 1.37 |
ENSDART00000169195
|
CT030188.1
|
|
| chr11_-_35171162 | 1.34 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr3_-_30488063 | 1.33 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr19_-_25119443 | 1.33 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr22_-_21897203 | 1.33 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr14_+_30340251 | 1.31 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr9_+_32178374 | 1.30 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr16_+_39159752 | 1.30 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr7_+_24528430 | 1.29 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr5_+_37406358 | 1.29 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr24_-_25166720 | 1.27 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr12_+_38563373 | 1.25 |
ENSDART00000134670
ENSDART00000193668 |
ttyh2
|
tweety family member 2 |
| chr8_-_9570511 | 1.25 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr16_-_41646164 | 1.24 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr21_-_36453594 | 1.24 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr15_-_31109760 | 1.23 |
ENSDART00000154254
|
ksr1b
|
kinase suppressor of ras 1b |
| chr7_+_40083601 | 1.23 |
ENSDART00000099046
|
zgc:112356
|
zgc:112356 |
| chr20_+_33987465 | 1.23 |
ENSDART00000061751
|
zp3a.2
|
zona pellucida glycoprotein 3a, tandem duplicate 2 |
| chr19_+_20201593 | 1.22 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr6_+_58280936 | 1.21 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr17_-_6641535 | 1.21 |
ENSDART00000154540
ENSDART00000180384 |
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr2_+_32846602 | 1.21 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr20_-_34028967 | 1.20 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr7_+_34794829 | 1.19 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr22_-_20695237 | 1.19 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr17_+_28628404 | 1.18 |
ENSDART00000032975
ENSDART00000143607 |
heatr5a
|
HEAT repeat containing 5a |
| chr22_-_10541372 | 1.17 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr16_-_17347727 | 1.17 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr17_-_2590222 | 1.16 |
ENSDART00000185711
|
CR759892.1
|
|
| chr19_-_11315224 | 1.15 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr11_+_31323746 | 1.15 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_+_7552699 | 1.14 |
ENSDART00000180788
ENSDART00000115058 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr19_-_34873566 | 1.14 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr17_+_25849332 | 1.13 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr17_+_50701748 | 1.13 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr12_-_18578218 | 1.13 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr8_+_42917515 | 1.13 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr12_-_18578432 | 1.13 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr5_+_25733774 | 1.11 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr16_+_35916371 | 1.11 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr14_+_34490445 | 1.09 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr20_+_27712714 | 1.08 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr2_-_21820697 | 1.08 |
ENSDART00000135230
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr13_+_5978809 | 1.07 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr13_-_4018888 | 1.05 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr17_-_8656155 | 1.03 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr19_-_34742440 | 1.02 |
ENSDART00000122625
ENSDART00000175621 |
elp2
|
elongator acetyltransferase complex subunit 2 |
| chr13_+_7665890 | 1.02 |
ENSDART00000046792
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr10_-_33297864 | 1.02 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr16_-_13680692 | 1.01 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr1_-_53407448 | 1.01 |
ENSDART00000160033
ENSDART00000172322 |
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr16_-_35427060 | 1.00 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr8_+_20488322 | 1.00 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr15_-_26931541 | 0.99 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr12_+_33320504 | 0.99 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr8_+_16676894 | 0.98 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr21_+_27513859 | 0.98 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr19_+_42086862 | 0.97 |
ENSDART00000151605
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr24_+_25794318 | 0.97 |
ENSDART00000143395
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr10_+_37181780 | 0.95 |
ENSDART00000187625
|
ksr1a
|
kinase suppressor of ras 1a |
| chr12_-_48188928 | 0.95 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr3_+_46271911 | 0.95 |
ENSDART00000186557
ENSDART00000113531 |
mkl2b
|
MKL/myocardin-like 2b |
| chr19_+_2685779 | 0.93 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr8_+_26874924 | 0.93 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr15_+_30310843 | 0.93 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr20_+_33991801 | 0.93 |
ENSDART00000061744
|
zp3a.1
|
zona pellucida glycoprotein 3a, tandem duplicate 1 |
| chr8_-_15129573 | 0.92 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr3_-_27647845 | 0.92 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr1_+_8521323 | 0.91 |
ENSDART00000121439
ENSDART00000103626 ENSDART00000141283 |
mief2
|
mitochondrial elongation factor 2 |
| chr22_+_14117078 | 0.91 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr8_-_50888806 | 0.91 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr4_+_9467049 | 0.91 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr18_+_26086803 | 0.90 |
ENSDART00000187911
|
znf710a
|
zinc finger protein 710a |
| chr13_-_35892051 | 0.90 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr20_-_28800999 | 0.90 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr15_+_23657051 | 0.89 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr11_+_31324335 | 0.89 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr16_+_54209504 | 0.88 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr21_+_4509483 | 0.88 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr23_-_40194732 | 0.88 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr22_+_23430688 | 0.88 |
ENSDART00000160457
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr11_-_26832685 | 0.87 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr2_-_38206034 | 0.86 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr8_-_29930821 | 0.86 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr23_-_14990865 | 0.85 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr14_+_30413758 | 0.85 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr24_-_19719240 | 0.85 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr3_-_32873641 | 0.84 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr17_+_1496107 | 0.84 |
ENSDART00000187804
|
LO018430.1
|
|
| chr9_-_31915423 | 0.84 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr1_-_23110740 | 0.83 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr21_-_36453417 | 0.83 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr9_+_43799829 | 0.83 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr13_-_35892243 | 0.82 |
ENSDART00000002750
ENSDART00000122810 ENSDART00000162399 |
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr15_+_34069746 | 0.82 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr12_+_23812530 | 0.81 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr12_-_8958011 | 0.81 |
ENSDART00000141212
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr3_+_35611625 | 0.80 |
ENSDART00000190995
|
traf7
|
TNF receptor-associated factor 7 |
| chr12_+_34953038 | 0.80 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr8_-_944055 | 0.79 |
ENSDART00000092773
|
mrps27
|
mitochondrial ribosomal protein S27 |
| chr16_-_42965192 | 0.79 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr23_+_4348479 | 0.79 |
ENSDART00000182425
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr19_+_15440841 | 0.79 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_+_6990970 | 0.79 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr11_+_31380495 | 0.79 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr16_-_24642814 | 0.78 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr20_-_51186524 | 0.78 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr22_-_10397600 | 0.78 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr23_+_19590006 | 0.78 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr19_-_5865766 | 0.78 |
ENSDART00000191007
|
LO018585.1
|
|
| chr13_-_33114933 | 0.78 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr5_-_1869982 | 0.78 |
ENSDART00000055878
|
rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr23_+_43684494 | 0.77 |
ENSDART00000149878
|
otud4
|
OTU deubiquitinase 4 |
| chr2_+_44972720 | 0.77 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr20_-_33512443 | 0.77 |
ENSDART00000061836
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr9_+_38457806 | 0.77 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr13_-_33227411 | 0.77 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr20_+_36812368 | 0.77 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr18_-_15532016 | 0.77 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr5_+_65086856 | 0.77 |
ENSDART00000169209
ENSDART00000162409 |
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr3_+_18807006 | 0.76 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr19_+_15441022 | 0.76 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr23_-_36441693 | 0.76 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr1_+_513986 | 0.76 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr15_+_15771418 | 0.75 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr15_-_17138640 | 0.75 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr2_-_57837838 | 0.74 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr21_+_4508959 | 0.74 |
ENSDART00000140432
ENSDART00000147187 ENSDART00000148910 |
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr5_+_65086668 | 0.73 |
ENSDART00000183746
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr7_+_48297842 | 0.73 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr9_+_16241656 | 0.73 |
ENSDART00000154326
|
si:ch211-261p9.4
|
si:ch211-261p9.4 |
| chr17_+_13664442 | 0.73 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr5_-_20921677 | 0.72 |
ENSDART00000158030
|
si:ch211-225b11.4
|
si:ch211-225b11.4 |
| chr19_+_28187480 | 0.72 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr23_+_20518504 | 0.71 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr8_-_51507144 | 0.70 |
ENSDART00000024882
ENSDART00000135166 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr15_-_43284021 | 0.70 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr15_+_15266025 | 0.70 |
ENSDART00000112974
ENSDART00000184450 |
c2cd3
|
C2 calcium-dependent domain containing 3 |
| chr12_+_21298317 | 0.70 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr24_+_22759451 | 0.70 |
ENSDART00000135392
|
si:dkey-7n6.2
|
si:dkey-7n6.2 |
| chr2_+_25315591 | 0.69 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr23_+_2914577 | 0.69 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr12_-_8958353 | 0.69 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr15_-_20745513 | 0.69 |
ENSDART00000187438
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr25_+_7299488 | 0.69 |
ENSDART00000184836
|
hmg20a
|
high mobility group 20A |
| chr2_-_26590628 | 0.69 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.3+nkx2.7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.1 | 3.4 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.5 | 1.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 3.6 | GO:0031670 | cellular response to nutrient(GO:0031670) cellular response to vitamin(GO:0071295) |
| 0.4 | 1.6 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.4 | 1.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 1.1 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.4 | 2.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.4 | 1.8 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.3 | 1.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 0.9 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.3 | 2.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 2.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 1.0 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.3 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 1.2 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 0.7 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.2 | 1.4 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 0.8 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.6 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.2 | 1.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 0.6 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.2 | 1.4 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.2 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.0 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 1.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.2 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.0 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 1.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 1.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.6 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.7 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 2.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 2.5 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.5 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.1 | 1.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.5 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.6 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 7.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 1.1 | GO:0048798 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.3 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 1.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 1.3 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.6 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 2.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.2 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 1.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 1.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 2.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.3 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.9 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.8 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 2.4 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.5 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.6 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 4.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 1.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 2.0 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.9 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.5 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 8.5 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 2.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.3 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 1.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.6 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 1.1 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.3 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.0 | 0.4 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 1.3 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.4 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.0 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.1 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.7 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.7 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.0 | 0.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 1.2 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.0 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.5 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 1.3 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.1 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.7 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 2.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 1.0 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 2.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 4.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 0.8 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 1.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 4.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.9 | GO:0098799 | mitochondrial outer membrane translocase complex(GO:0005742) outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 2.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 3.1 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.7 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 0.4 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 2.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 3.5 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.6 | 4.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.5 | 1.6 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.5 | 1.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.0 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.3 | 0.9 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.3 | 2.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 3.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.3 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 1.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 1.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 2.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 2.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 1.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 2.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.5 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.5 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 4.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.6 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 3.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 3.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 7.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 1.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.7 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 4.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.5 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 2.0 | GO:0042802 | identical protein binding(GO:0042802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 0.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 3.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 1.9 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.2 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |