Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for nkx2.2b
Z-value: 0.94
Transcription factors associated with nkx2.2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2b
|
ENSDARG00000101549 | NK2 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2b | dr11_v1_chr20_+_48782068_48782068 | -0.50 | 3.4e-02 | Click! |
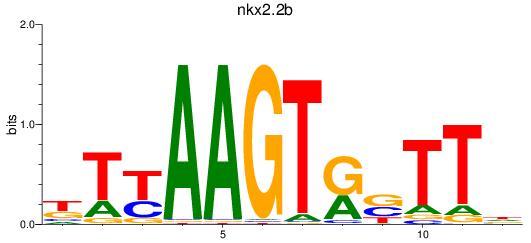
Activity profile of nkx2.2b motif
Sorted Z-values of nkx2.2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_47710434 | 1.95 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr4_-_837768 | 1.93 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr4_-_20081621 | 1.74 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr22_-_17631675 | 1.73 |
ENSDART00000132565
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr24_-_33308045 | 1.72 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr10_+_41945890 | 1.67 |
ENSDART00000063013
ENSDART00000128313 |
tmem120b
|
transmembrane protein 120B |
| chr11_+_42422371 | 1.67 |
ENSDART00000163780
|
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr17_+_12075805 | 1.60 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr6_-_49547680 | 1.54 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr7_-_71585065 | 1.49 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr6_-_19341184 | 1.46 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr12_+_33320884 | 1.43 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr6_+_10338554 | 1.43 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr8_+_52442622 | 1.42 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr13_-_33317323 | 1.41 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr13_+_8840772 | 1.41 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr10_-_32877348 | 1.37 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr18_-_35407695 | 1.34 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr23_+_36730713 | 1.32 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr11_-_25257595 | 1.32 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr16_-_8132742 | 1.29 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr4_-_20108833 | 1.29 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr14_+_26759332 | 1.28 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr2_-_42552666 | 1.28 |
ENSDART00000141399
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr24_-_32522587 | 1.27 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr2_-_10877228 | 1.27 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr11_+_37638873 | 1.26 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr12_-_4301234 | 1.26 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr16_-_4610255 | 1.25 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr13_+_33368140 | 1.25 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr16_+_40340523 | 1.24 |
ENSDART00000102571
|
mettl6
|
methyltransferase like 6 |
| chr1_-_40341306 | 1.23 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr20_-_32045057 | 1.21 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr6_-_49537646 | 1.21 |
ENSDART00000180438
|
FO704848.1
|
|
| chr16_-_22585289 | 1.20 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr8_-_1698155 | 1.19 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr1_-_43905252 | 1.19 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr7_-_26518086 | 1.18 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr22_+_883678 | 1.15 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr24_-_26518972 | 1.15 |
ENSDART00000097792
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr6_-_8465656 | 1.13 |
ENSDART00000178887
|
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr20_+_25712276 | 1.12 |
ENSDART00000121585
ENSDART00000185772 |
cep135
|
centrosomal protein 135 |
| chr14_+_30398546 | 1.11 |
ENSDART00000053925
|
mtmr7a
|
myotubularin related protein 7a |
| chr6_+_21227621 | 1.10 |
ENSDART00000193583
|
prkca
|
protein kinase C, alpha |
| chr25_-_998096 | 1.09 |
ENSDART00000164082
|
znf609a
|
zinc finger protein 609a |
| chr11_+_42422638 | 1.08 |
ENSDART00000042599
ENSDART00000181175 |
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr1_-_55248496 | 1.08 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr1_+_30723380 | 1.07 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr12_-_25150239 | 1.07 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr1_-_354115 | 1.07 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr8_+_3431671 | 1.06 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr9_-_14273652 | 1.04 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr10_+_20392083 | 1.03 |
ENSDART00000166867
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr4_-_12477224 | 1.03 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr14_+_22132896 | 0.99 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr2_-_21786826 | 0.98 |
ENSDART00000016208
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr15_-_37589600 | 0.98 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr24_+_41989108 | 0.97 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr4_-_5108844 | 0.95 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr7_-_64770456 | 0.95 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr20_+_53368611 | 0.94 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr5_-_57204352 | 0.94 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr20_-_2298970 | 0.93 |
ENSDART00000136067
|
EPB41L2
|
si:ch73-18b11.1 |
| chr11_+_18873619 | 0.93 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr2_-_10943093 | 0.93 |
ENSDART00000148999
|
ssbp3a
|
single stranded DNA binding protein 3a |
| chr18_+_26428829 | 0.92 |
ENSDART00000190779
ENSDART00000193226 ENSDART00000110746 |
blm
|
Bloom syndrome, RecQ helicase-like |
| chr19_-_5058908 | 0.92 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr6_+_1724889 | 0.92 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr13_+_33368503 | 0.91 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr12_-_1031970 | 0.91 |
ENSDART00000105292
|
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr5_+_16117871 | 0.91 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr14_+_22132388 | 0.91 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr3_+_39579393 | 0.91 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr13_+_36595618 | 0.91 |
ENSDART00000022684
|
cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr18_-_27316599 | 0.90 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr22_+_28818291 | 0.90 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr1_-_47122058 | 0.90 |
ENSDART00000159925
ENSDART00000101143 ENSDART00000176803 |
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr12_-_22400999 | 0.90 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr22_-_21676364 | 0.89 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr14_-_26392475 | 0.88 |
ENSDART00000170614
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr22_-_20924564 | 0.87 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr20_+_46620374 | 0.87 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr17_+_19481049 | 0.86 |
ENSDART00000024194
|
kif11
|
kinesin family member 11 |
| chr13_+_15581270 | 0.86 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr4_+_16784553 | 0.85 |
ENSDART00000134054
|
si:dkey-13i19.8
|
si:dkey-13i19.8 |
| chr16_-_31284922 | 0.85 |
ENSDART00000142638
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr16_+_20056030 | 0.85 |
ENSDART00000027020
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr3_+_18807524 | 0.84 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr12_-_3705862 | 0.83 |
ENSDART00000193864
ENSDART00000185857 |
CABZ01069040.1
|
|
| chr2_-_32387441 | 0.82 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr3_-_26806032 | 0.82 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr22_+_35131890 | 0.82 |
ENSDART00000003303
ENSDART00000130581 |
rnf13
|
ring finger protein 13 |
| chr3_-_34136778 | 0.81 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr4_+_5334202 | 0.79 |
ENSDART00000150409
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr23_+_4226341 | 0.78 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr5_-_33886495 | 0.78 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr17_-_29312506 | 0.78 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr4_+_5196469 | 0.77 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr22_+_835728 | 0.77 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr24_+_24726956 | 0.77 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr13_-_32726178 | 0.76 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr24_+_10039165 | 0.76 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr20_-_53963515 | 0.76 |
ENSDART00000110252
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr21_+_43178831 | 0.75 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr19_+_24039830 | 0.75 |
ENSDART00000100422
|
rit1
|
Ras-like without CAAX 1 |
| chr5_+_3927989 | 0.75 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr14_+_45608576 | 0.75 |
ENSDART00000173248
|
si:ch211-276i12.11
|
si:ch211-276i12.11 |
| chr10_-_7702029 | 0.74 |
ENSDART00000018017
|
gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr17_+_6538733 | 0.73 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr21_+_19547806 | 0.73 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr19_-_5058515 | 0.71 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr24_-_31332087 | 0.71 |
ENSDART00000161179
|
abcd3a
|
ATP-binding cassette, sub-family D (ALD), member 3a |
| chr16_+_39242339 | 0.71 |
ENSDART00000102510
|
zgc:77056
|
zgc:77056 |
| chr8_-_49728590 | 0.71 |
ENSDART00000135714
ENSDART00000138810 ENSDART00000098319 |
gkap1
|
G kinase anchoring protein 1 |
| chr19_+_30867845 | 0.70 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr15_+_24746444 | 0.70 |
ENSDART00000032306
|
ywhae1
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 |
| chr3_-_26805455 | 0.70 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr12_+_19138452 | 0.69 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr20_+_2739531 | 0.69 |
ENSDART00000152269
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr12_+_33361948 | 0.68 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr17_+_22577472 | 0.68 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr2_-_22530969 | 0.67 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr20_-_28698172 | 0.67 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr11_-_11961706 | 0.67 |
ENSDART00000115249
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr2_-_58183499 | 0.67 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr4_+_5333988 | 0.67 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr16_-_25400257 | 0.66 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr22_+_11775269 | 0.66 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr2_+_2967255 | 0.65 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr7_+_37742299 | 0.65 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr10_+_585719 | 0.64 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr19_+_7173613 | 0.64 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr7_+_32901658 | 0.63 |
ENSDART00000115420
|
ano9b
|
anoctamin 9b |
| chr3_-_11878490 | 0.63 |
ENSDART00000129961
|
coro7
|
coronin 7 |
| chr20_+_2739804 | 0.63 |
ENSDART00000152655
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr4_-_16876281 | 0.63 |
ENSDART00000016690
ENSDART00000044005 ENSDART00000042874 ENSDART00000125762 ENSDART00000185974 |
tmpoa
|
thymopoietin a |
| chr5_+_5406503 | 0.62 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr16_+_14249546 | 0.62 |
ENSDART00000059967
|
polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr10_-_6494070 | 0.61 |
ENSDART00000171320
|
dcp2
|
decapping mRNA 2 |
| chr14_+_23717165 | 0.61 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr15_-_20731638 | 0.61 |
ENSDART00000170616
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr4_+_73051901 | 0.61 |
ENSDART00000174219
|
zgc:152938
|
zgc:152938 |
| chr11_+_5681762 | 0.61 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_+_25157374 | 0.60 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr15_-_20731297 | 0.60 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr2_+_51783120 | 0.59 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr5_+_43782267 | 0.59 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr5_+_31965845 | 0.59 |
ENSDART00000112968
|
myo1hb
|
myosin IHb |
| chr11_+_3308656 | 0.58 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr7_+_39054478 | 0.58 |
ENSDART00000173825
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr5_+_27137473 | 0.58 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr6_+_18544791 | 0.58 |
ENSDART00000167463
ENSDART00000169599 |
atad5b
|
ATPase family, AAA domain containing 5b |
| chr2_-_15324837 | 0.57 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr18_-_22735002 | 0.57 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr6_+_55212928 | 0.57 |
ENSDART00000083668
|
manbal
|
mannosidase beta like |
| chr14_+_1170968 | 0.56 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr5_+_30596632 | 0.56 |
ENSDART00000051414
|
hinfp
|
histone H4 transcription factor |
| chr12_-_28794957 | 0.56 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr6_-_42377307 | 0.55 |
ENSDART00000129302
|
emc3
|
ER membrane protein complex subunit 3 |
| chr17_-_33412868 | 0.55 |
ENSDART00000187521
|
BX323819.1
|
|
| chr6_-_36795111 | 0.54 |
ENSDART00000160669
ENSDART00000104256 ENSDART00000187751 ENSDART00000161928 ENSDART00000183264 |
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr14_-_21097574 | 0.54 |
ENSDART00000186803
|
rnf20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr5_+_30596477 | 0.52 |
ENSDART00000124487
|
hinfp
|
histone H4 transcription factor |
| chr13_-_25819825 | 0.52 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr8_-_48668111 | 0.51 |
ENSDART00000047134
|
clcn6
|
chloride channel 6 |
| chr5_+_18012154 | 0.51 |
ENSDART00000139431
ENSDART00000048859 |
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr2_-_37280617 | 0.51 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr14_+_29550323 | 0.50 |
ENSDART00000157943
|
TENM2
|
si:dkey-34l15.2 |
| chr17_-_31819837 | 0.50 |
ENSDART00000160281
|
abraxas2b
|
abraxas 2b, BRISC complex subunit |
| chr19_+_26922780 | 0.49 |
ENSDART00000187396
ENSDART00000188978 |
nelfe
|
negative elongation factor complex member E |
| chr13_-_865193 | 0.49 |
ENSDART00000187053
|
AL929536.7
|
|
| chr20_+_9128256 | 0.49 |
ENSDART00000163883
ENSDART00000183072 ENSDART00000187276 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr7_+_30725473 | 0.49 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr25_+_2645488 | 0.49 |
ENSDART00000113032
|
adck2
|
aarF domain containing kinase 2 |
| chr23_+_43770149 | 0.49 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr3_+_19685873 | 0.48 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr9_-_14683574 | 0.48 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr13_-_30996072 | 0.48 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr21_+_15603597 | 0.47 |
ENSDART00000138626
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr5_-_55914268 | 0.47 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr14_+_14043793 | 0.47 |
ENSDART00000164376
|
rraga
|
Ras-related GTP binding A |
| chr21_+_76739 | 0.47 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr12_-_10476448 | 0.46 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr17_+_50074372 | 0.45 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr12_+_33484458 | 0.45 |
ENSDART00000000069
|
slc9a3r1a
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1a |
| chr13_-_22699024 | 0.45 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr11_+_18873113 | 0.44 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr3_+_19216567 | 0.43 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr6_+_40951227 | 0.43 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr19_+_26923274 | 0.43 |
ENSDART00000148439
ENSDART00000148877 |
nelfe
|
negative elongation factor complex member E |
| chr5_+_1965296 | 0.43 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr20_-_28884800 | 0.42 |
ENSDART00000134564
ENSDART00000132127 ENSDART00000135506 ENSDART00000075515 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr7_+_31132588 | 0.42 |
ENSDART00000173702
|
tjp1a
|
tight junction protein 1a |
| chr8_-_51954562 | 0.41 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
| chr19_+_42231431 | 0.41 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr12_+_36109507 | 0.40 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr24_+_35183595 | 0.40 |
ENSDART00000075142
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr7_+_7511914 | 0.40 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr8_+_13368150 | 0.39 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr6_-_31739709 | 0.39 |
ENSDART00000087964
|
cachd1
|
cache domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.4 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.4 | 1.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.9 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 1.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 0.7 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 1.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 1.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.2 | 1.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.5 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.2 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.2 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.9 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.9 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.7 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.6 | GO:0090133 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 1.0 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.9 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.6 | GO:0031284 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.5 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 0.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 1.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 2.0 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 1.4 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.9 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:0045943 | regulation of transcription from RNA polymerase I promoter(GO:0006356) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.6 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 1.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.5 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 1.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.4 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 1.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.8 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 1.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.8 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.4 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.9 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.7 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.9 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.3 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.9 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.4 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.5 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.6 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0033135 | regulation of peptidyl-serine phosphorylation(GO:0033135) positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.4 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.9 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 2.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.3 | 1.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.5 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 2.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 4.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.3 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.1 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 3.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 2.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) INO80-type complex(GO:0097346) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 2.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.3 | 1.5 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 0.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.5 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.6 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.6 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.9 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.9 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.6 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.5 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.8 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |