Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
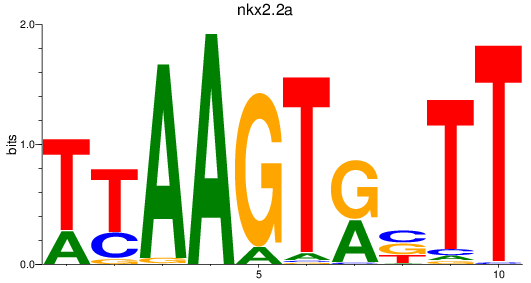
Results for nkx2.2a
Z-value: 0.24
Transcription factors associated with nkx2.2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2a
|
ENSDARG00000053298 | NK2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2a | dr11_v1_chr17_-_42213822_42213822 | 0.22 | 3.8e-01 | Click! |
Activity profile of nkx2.2a motif
Sorted Z-values of nkx2.2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_33308045 | 0.25 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr7_+_22792895 | 0.25 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr19_+_24039830 | 0.22 |
ENSDART00000100422
|
rit1
|
Ras-like without CAAX 1 |
| chr2_+_51818039 | 0.20 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr11_+_42422371 | 0.20 |
ENSDART00000163780
|
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chrM_+_12897 | 0.19 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr22_-_8006342 | 0.18 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr2_-_38225388 | 0.18 |
ENSDART00000146485
ENSDART00000128043 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr13_+_47710434 | 0.18 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr5_-_54481692 | 0.17 |
ENSDART00000165719
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr20_-_9194257 | 0.17 |
ENSDART00000133012
|
ylpm1
|
YLP motif containing 1 |
| chr16_-_54919260 | 0.17 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr10_+_44984946 | 0.16 |
ENSDART00000182520
|
h2afvb
|
H2A histone family, member Vb |
| chr1_-_23294753 | 0.16 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr11_-_11518469 | 0.16 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr19_-_27827744 | 0.16 |
ENSDART00000181620
|
papd7
|
PAP associated domain containing 7 |
| chr4_-_4570475 | 0.16 |
ENSDART00000184955
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr10_-_32851847 | 0.15 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr7_+_22792132 | 0.15 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr17_+_19481049 | 0.14 |
ENSDART00000024194
|
kif11
|
kinesin family member 11 |
| chr10_-_10018120 | 0.14 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr6_-_8465656 | 0.14 |
ENSDART00000178887
|
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr20_+_25712276 | 0.14 |
ENSDART00000121585
ENSDART00000185772 |
cep135
|
centrosomal protein 135 |
| chr4_-_72609735 | 0.14 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr14_+_46342882 | 0.14 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr1_-_40341306 | 0.14 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr15_+_15266025 | 0.14 |
ENSDART00000112974
ENSDART00000184450 |
c2cd3
|
C2 calcium-dependent domain containing 3 |
| chr6_+_10338554 | 0.14 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr16_-_33105677 | 0.14 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr19_+_7575341 | 0.14 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr22_+_23430688 | 0.14 |
ENSDART00000160457
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr21_-_21790372 | 0.13 |
ENSDART00000151094
|
xrra1
|
X-ray radiation resistance associated 1 |
| chr16_-_4610255 | 0.13 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr13_-_38730267 | 0.13 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr24_+_17069420 | 0.13 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr2_+_31437547 | 0.13 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr9_+_28140089 | 0.13 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr7_+_41887429 | 0.13 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr14_-_48786708 | 0.13 |
ENSDART00000169730
|
si:ch211-199b20.3
|
si:ch211-199b20.3 |
| chr11_+_34522554 | 0.13 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr21_+_19547806 | 0.13 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr11_+_42422638 | 0.13 |
ENSDART00000042599
ENSDART00000181175 |
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr16_-_39267185 | 0.13 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr10_+_41945890 | 0.13 |
ENSDART00000063013
ENSDART00000128313 |
tmem120b
|
transmembrane protein 120B |
| chr11_+_25064519 | 0.12 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr4_-_27129697 | 0.12 |
ENSDART00000131240
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr5_+_41143563 | 0.12 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr24_-_2423791 | 0.12 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr10_+_36441124 | 0.12 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr8_-_1698155 | 0.12 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr16_-_31284922 | 0.12 |
ENSDART00000142638
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr10_-_7988396 | 0.12 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr2_-_32387441 | 0.12 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr22_+_28818291 | 0.12 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr3_+_31662126 | 0.12 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr23_-_270847 | 0.12 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr2_+_2967255 | 0.11 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr16_-_47427016 | 0.11 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr22_-_34979139 | 0.11 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr18_+_26428829 | 0.11 |
ENSDART00000190779
ENSDART00000193226 ENSDART00000110746 |
blm
|
Bloom syndrome, RecQ helicase-like |
| chr8_-_11016766 | 0.11 |
ENSDART00000064066
|
bcas2
|
BCAS2, pre-mRNA processing factor |
| chr8_-_49764156 | 0.11 |
ENSDART00000177589
ENSDART00000189142 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr6_+_11438972 | 0.11 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr11_+_3308656 | 0.11 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr2_-_32262287 | 0.11 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr12_-_11349899 | 0.11 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr11_+_12811906 | 0.11 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr1_-_681116 | 0.10 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr17_+_30205258 | 0.10 |
ENSDART00000076596
ENSDART00000153795 |
spata17
|
spermatogenesis associated 17 |
| chr6_+_21227621 | 0.10 |
ENSDART00000193583
|
prkca
|
protein kinase C, alpha |
| chr16_-_22585289 | 0.10 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr17_+_21887823 | 0.10 |
ENSDART00000131929
ENSDART00000165192 |
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr5_+_43782267 | 0.10 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr1_-_9858508 | 0.10 |
ENSDART00000147904
|
mad1l1
|
mitotic arrest deficient 1 like 1 |
| chr19_+_29798064 | 0.10 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr13_-_33317323 | 0.10 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr23_+_19813677 | 0.10 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr1_-_625875 | 0.10 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr7_-_64770456 | 0.10 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr13_-_865193 | 0.10 |
ENSDART00000187053
|
AL929536.7
|
|
| chr20_-_53963515 | 0.10 |
ENSDART00000110252
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr2_+_50477779 | 0.10 |
ENSDART00000122716
|
CABZ01067973.1
|
|
| chr3_+_39579393 | 0.10 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr12_+_33361948 | 0.10 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr11_-_30636163 | 0.09 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr18_-_17145111 | 0.09 |
ENSDART00000090446
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr3_+_16668609 | 0.09 |
ENSDART00000185646
|
zgc:55558
|
zgc:55558 |
| chr25_-_5119162 | 0.09 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr11_+_18873113 | 0.09 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr23_+_6795709 | 0.09 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr3_+_31093455 | 0.09 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr19_+_34230108 | 0.09 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr4_-_12914163 | 0.09 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr6_+_1724889 | 0.09 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr23_-_36934944 | 0.09 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr7_-_40122139 | 0.09 |
ENSDART00000173982
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr22_+_17248145 | 0.09 |
ENSDART00000136908
|
axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr24_+_5935377 | 0.09 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr3_-_26805455 | 0.08 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr22_+_38049130 | 0.08 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr14_-_49133426 | 0.08 |
ENSDART00000042421
|
ppp2ca
|
protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr17_+_50074372 | 0.08 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr23_-_14990865 | 0.08 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr3_-_31875138 | 0.08 |
ENSDART00000155298
|
limd2
|
LIM domain containing 2 |
| chr16_-_8132742 | 0.08 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr9_+_35016201 | 0.08 |
ENSDART00000182404
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr13_+_33368140 | 0.08 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr19_+_14352332 | 0.08 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr13_-_22699024 | 0.08 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr3_-_26806032 | 0.08 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr1_-_54622227 | 0.08 |
ENSDART00000049010
|
tekt4
|
tektin 4 |
| chr15_-_28569786 | 0.08 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr12_+_33320884 | 0.08 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr1_+_37196106 | 0.08 |
ENSDART00000008756
ENSDART00000157503 ENSDART00000162971 ENSDART00000191004 ENSDART00000078206 ENSDART00000045111 |
dclk2a
|
doublecortin-like kinase 2a |
| chr21_+_30662263 | 0.08 |
ENSDART00000154758
ENSDART00000138664 |
clcn5a
|
chloride channel, voltage-sensitive 5a |
| chr14_-_858985 | 0.08 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr14_-_25956804 | 0.08 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr5_-_38777852 | 0.08 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr7_+_20109968 | 0.08 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr16_+_32729223 | 0.08 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr8_+_3431671 | 0.08 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr5_-_23473312 | 0.08 |
ENSDART00000140310
|
si:dkeyp-20g2.3
|
si:dkeyp-20g2.3 |
| chr7_+_42328578 | 0.08 |
ENSDART00000149082
|
phkb
|
phosphorylase kinase, beta |
| chr7_-_7766920 | 0.08 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr24_+_15655069 | 0.08 |
ENSDART00000042943
ENSDART00000181296 |
fbxo15
|
F-box protein 15 |
| chr17_+_27723490 | 0.08 |
ENSDART00000123588
ENSDART00000170462 ENSDART00000169708 |
qkia
|
QKI, KH domain containing, RNA binding a |
| chr5_-_10082244 | 0.07 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr23_+_6795531 | 0.07 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr16_+_17763848 | 0.07 |
ENSDART00000149408
ENSDART00000148878 |
them4
|
thioesterase superfamily member 4 |
| chr9_+_34950942 | 0.07 |
ENSDART00000077800
|
tfdp1a
|
transcription factor Dp-1, a |
| chr24_-_7587401 | 0.07 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr9_-_33781845 | 0.07 |
ENSDART00000008737
|
efhc2
|
EF-hand domain (C-terminal) containing 2 |
| chr24_+_41989108 | 0.07 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr15_-_20731297 | 0.07 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr5_-_19444930 | 0.07 |
ENSDART00000136259
ENSDART00000188499 |
kctd10
|
potassium channel tetramerization domain containing 10 |
| chr12_-_5188413 | 0.07 |
ENSDART00000161988
|
fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr24_-_26518972 | 0.07 |
ENSDART00000097792
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr23_-_17451264 | 0.07 |
ENSDART00000190748
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr24_+_10039165 | 0.07 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr9_-_15208129 | 0.07 |
ENSDART00000137043
ENSDART00000131512 |
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr15_+_31526225 | 0.07 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr6_+_9793791 | 0.07 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr22_-_21676364 | 0.07 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr12_-_25150239 | 0.07 |
ENSDART00000038415
ENSDART00000135368 |
rhoq
|
ras homolog family member Q |
| chr2_-_10877228 | 0.07 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr7_+_13527971 | 0.07 |
ENSDART00000020542
|
plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr14_+_23717165 | 0.07 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr12_-_3705862 | 0.07 |
ENSDART00000193864
ENSDART00000185857 |
CABZ01069040.1
|
|
| chr7_+_71586485 | 0.07 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr5_+_5406503 | 0.07 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr11_-_11961706 | 0.07 |
ENSDART00000115249
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr2_+_33051730 | 0.06 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr14_-_38889840 | 0.06 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr23_+_20431388 | 0.06 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr22_-_16270462 | 0.06 |
ENSDART00000105681
|
cdc14ab
|
cell division cycle 14Ab |
| chr20_-_54245256 | 0.06 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr25_+_8407892 | 0.06 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr3_-_15487111 | 0.06 |
ENSDART00000011320
|
nfatc2ip
|
nuclear factor of activated T cells 2 interacting protein |
| chr13_-_31164749 | 0.06 |
ENSDART00000049180
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr20_+_812012 | 0.06 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr10_-_5135788 | 0.06 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr14_+_14043793 | 0.06 |
ENSDART00000164376
|
rraga
|
Ras-related GTP binding A |
| chr3_+_46315016 | 0.06 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr23_+_36730713 | 0.06 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr22_-_15562933 | 0.06 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr19_-_10656667 | 0.06 |
ENSDART00000081379
ENSDART00000151456 ENSDART00000143271 ENSDART00000182126 |
olah
|
oleoyl-ACP hydrolase |
| chr2_-_7185460 | 0.06 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr20_-_33507458 | 0.06 |
ENSDART00000140287
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr15_-_20731638 | 0.06 |
ENSDART00000170616
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr19_-_32940040 | 0.06 |
ENSDART00000179947
|
azin1b
|
antizyme inhibitor 1b |
| chr7_-_69636502 | 0.06 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr13_+_33368503 | 0.06 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr4_+_37992753 | 0.06 |
ENSDART00000193890
|
CR759843.2
|
|
| chr24_-_37272116 | 0.06 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr25_-_10615504 | 0.06 |
ENSDART00000156797
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr17_+_27545183 | 0.06 |
ENSDART00000129392
|
pacrg
|
PARK2 co-regulated |
| chr1_-_58601636 | 0.06 |
ENSDART00000141143
|
si:ch73-236c18.8
|
si:ch73-236c18.8 |
| chr3_-_34136778 | 0.06 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr9_+_43799829 | 0.06 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr21_-_27221535 | 0.05 |
ENSDART00000138266
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr5_+_54497730 | 0.05 |
ENSDART00000157722
|
tmem203
|
transmembrane protein 203 |
| chr25_-_30047477 | 0.05 |
ENSDART00000164859
|
CR759810.1
|
|
| chr19_+_24896409 | 0.05 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr1_-_1894722 | 0.05 |
ENSDART00000165669
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr1_-_51261420 | 0.05 |
ENSDART00000168685
|
kif16ba
|
kinesin family member 16Ba |
| chr13_-_18863680 | 0.05 |
ENSDART00000109277
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr25_+_3358701 | 0.05 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr15_-_23529945 | 0.05 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr12_-_8817999 | 0.05 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr12_+_34770531 | 0.05 |
ENSDART00000153320
|
slc38a10
|
solute carrier family 38, member 10 |
| chr12_-_4424424 | 0.05 |
ENSDART00000152314
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr15_+_6786757 | 0.05 |
ENSDART00000013278
|
sirt2
|
sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) |
| chr6_-_41085692 | 0.05 |
ENSDART00000181463
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr19_+_26923274 | 0.05 |
ENSDART00000148439
ENSDART00000148877 |
nelfe
|
negative elongation factor complex member E |
| chr6_-_36795111 | 0.05 |
ENSDART00000160669
ENSDART00000104256 ENSDART00000187751 ENSDART00000161928 ENSDART00000183264 |
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr14_-_25985698 | 0.05 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr12_-_24812403 | 0.05 |
ENSDART00000185517
|
foxn2b
|
forkhead box N2b |
| chr11_+_37638873 | 0.05 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr16_-_25400257 | 0.05 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr16_-_41465542 | 0.05 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.1 | GO:0072156 | proximal/distal pattern formation involved in nephron development(GO:0072047) distal tubule morphogenesis(GO:0072156) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.1 | GO:1904867 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0030828 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.0 | GO:0033335 | anal fin development(GO:0033335) |
| 0.0 | 0.1 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.0 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |