Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
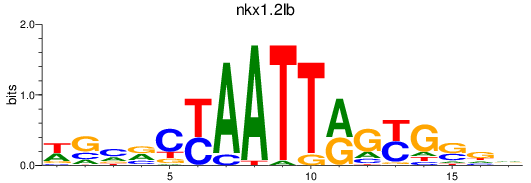
Results for nkx1.2lb
Z-value: 0.36
Transcription factors associated with nkx1.2lb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx1.2lb
|
ENSDARG00000099427 | NK1 transcription factor related 2-like,b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx1.2lb | dr11_v1_chr14_+_15064870_15064870 | -0.36 | 1.4e-01 | Click! |
Activity profile of nkx1.2lb motif
Sorted Z-values of nkx1.2lb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23780334 | 0.79 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr10_-_34002185 | 0.74 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr2_+_6253246 | 0.72 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr20_-_14114078 | 0.65 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr20_+_14114258 | 0.58 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr19_-_18313303 | 0.44 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr17_+_24821627 | 0.38 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr25_-_32869794 | 0.35 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr4_+_11458078 | 0.34 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr9_-_50000144 | 0.32 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr1_+_55758257 | 0.27 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr13_-_9236905 | 0.26 |
ENSDART00000142597
ENSDART00000147176 ENSDART00000181670 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-33c12.12 si:dkey-112g5.12 |
| chr13_-_9213207 | 0.24 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr20_+_36812368 | 0.22 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr11_-_16152400 | 0.21 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr21_+_4410733 | 0.21 |
ENSDART00000136831
ENSDART00000131612 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr21_+_4328348 | 0.21 |
ENSDART00000131658
ENSDART00000143799 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr8_+_18011522 | 0.18 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr1_-_48933 | 0.18 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr23_+_29358188 | 0.18 |
ENSDART00000189242
|
tardbpl
|
TAR DNA binding protein, like |
| chr2_+_10007113 | 0.18 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr2_+_10006839 | 0.17 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr7_-_64971839 | 0.17 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr13_-_17723417 | 0.17 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr17_+_16429826 | 0.17 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr20_-_37813863 | 0.17 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr13_+_35955562 | 0.17 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr13_+_9521629 | 0.16 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr21_+_4348600 | 0.16 |
ENSDART00000138463
|
HTRA2 (1 of many)
|
si:dkey-84o3.2 |
| chr13_-_9159852 | 0.16 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr13_-_9257728 | 0.15 |
ENSDART00000145985
ENSDART00000169991 ENSDART00000132764 |
HTRA2 (1 of many)
|
zgc:173425 |
| chr5_-_33298352 | 0.15 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr2_+_50608099 | 0.15 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr13_+_12761707 | 0.15 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr13_+_9468535 | 0.15 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr7_+_20384408 | 0.15 |
ENSDART00000173809
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr6_+_23026714 | 0.14 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr21_+_4429001 | 0.14 |
ENSDART00000134520
|
HTRA2 (1 of many)
|
si:dkey-84o3.6 |
| chr13_-_9111927 | 0.14 |
ENSDART00000133815
ENSDART00000109783 ENSDART00000142540 ENSDART00000192613 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-19p15.4 si:dkey-112g5.12 |
| chr13_-_6279684 | 0.13 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr3_-_40836081 | 0.13 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr12_-_19119176 | 0.12 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr20_+_43925266 | 0.12 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr13_-_9184132 | 0.12 |
ENSDART00000135289
ENSDART00000136309 ENSDART00000132630 |
HTRA2 (1 of many)
|
si:dkey-33c12.10 |
| chr13_-_9045879 | 0.11 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr7_-_40630698 | 0.11 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr7_+_20383841 | 0.11 |
ENSDART00000052906
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr7_-_59123066 | 0.11 |
ENSDART00000175438
|
dennd4c
|
DENN/MADD domain containing 4C |
| chr13_-_9070754 | 0.10 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr12_-_4249000 | 0.10 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr3_+_17537352 | 0.10 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr1_-_8140763 | 0.09 |
ENSDART00000091508
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr13_-_30142087 | 0.09 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr13_+_9496748 | 0.08 |
ENSDART00000147050
ENSDART00000102120 |
CR848040.4
|
|
| chr17_+_21902058 | 0.08 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr21_+_4368735 | 0.07 |
ENSDART00000182817
|
CT027677.1
|
|
| chr16_+_4078240 | 0.07 |
ENSDART00000160890
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr17_-_53329704 | 0.05 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr10_-_105100 | 0.05 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr17_-_15189397 | 0.05 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr6_-_12275836 | 0.05 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr7_+_24881680 | 0.04 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr9_-_35334642 | 0.04 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr1_-_44812014 | 0.04 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr14_+_918287 | 0.03 |
ENSDART00000167066
|
CU462913.1
|
|
| chr20_-_16972351 | 0.03 |
ENSDART00000148312
ENSDART00000186702 |
si:ch73-74h11.1
|
si:ch73-74h11.1 |
| chr23_-_35064785 | 0.03 |
ENSDART00000172240
|
BX294434.1
|
|
| chr24_-_32582378 | 0.03 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr11_+_12811906 | 0.03 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr23_+_26946744 | 0.02 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr9_+_43799829 | 0.02 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr25_+_36312459 | 0.02 |
ENSDART00000182484
|
CR354435.5
|
|
| chr6_+_4255319 | 0.02 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr7_+_54642005 | 0.02 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr21_-_33995710 | 0.02 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr12_-_43685802 | 0.02 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr16_+_32051572 | 0.02 |
ENSDART00000039109
|
leng1
|
leukocyte receptor cluster (LRC) member 1 |
| chr5_-_22052852 | 0.02 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr8_-_20243389 | 0.02 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr21_-_21570134 | 0.02 |
ENSDART00000174388
|
or133-4
|
odorant receptor, family H, subfamily 133, member 4 |
| chr8_-_29658171 | 0.02 |
ENSDART00000147755
|
mpeg1.3
|
macrophage expressed 1, tandem duplicate 3 |
| chr23_-_1348933 | 0.01 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr8_+_22472584 | 0.01 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr14_-_30945515 | 0.01 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr5_-_51619742 | 0.01 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr7_+_34453185 | 0.01 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr12_+_3078221 | 0.01 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr21_-_15200556 | 0.01 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr23_-_44965582 | 0.01 |
ENSDART00000163367
|
tfr2
|
transferrin receptor 2 |
| chr4_-_42408339 | 0.00 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr23_-_24263474 | 0.00 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr1_-_50791280 | 0.00 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr25_+_20272145 | 0.00 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr15_-_25518084 | 0.00 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx1.2lb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.4 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.1 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |