Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for nfkb1_rela
Z-value: 0.70
Transcription factors associated with nfkb1_rela
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
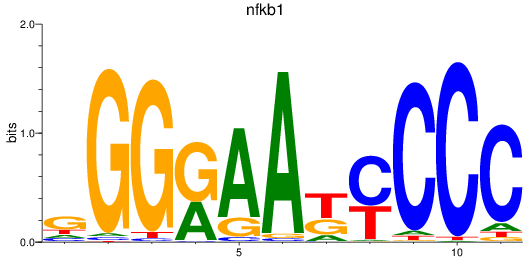
nfkb1
|
ENSDARG00000105261 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
|
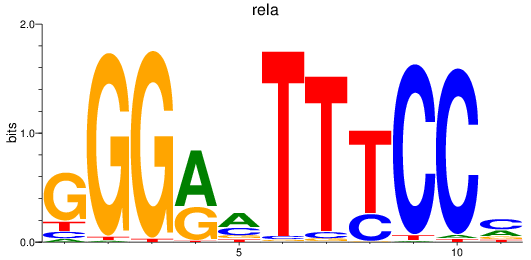
rela
|
ENSDARG00000098696 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfkb1 | dr11_v1_chr14_-_49859747_49859823 | 0.98 | 9.9e-13 | Click! |
| rela | dr11_v1_chr7_-_804515_804515 | 0.22 | 3.8e-01 | Click! |
Activity profile of nfkb1_rela motif
Sorted Z-values of nfkb1_rela motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_51001985 | 1.32 |
ENSDART00000074638
|
ankrd31
|
ankyrin repeat domain 31 |
| chr4_-_12795436 | 1.31 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr1_-_37377509 | 1.29 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr4_+_3389866 | 1.18 |
ENSDART00000153834
|
sypl1
|
synaptophysin-like 1 |
| chr5_-_29780752 | 1.17 |
ENSDART00000137400
ENSDART00000145021 |
cfap77
|
cilia and flagella associated protein 77 |
| chr14_-_49859747 | 1.17 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr12_+_7445595 | 1.14 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr12_+_26471712 | 1.08 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr14_+_3038473 | 1.06 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr23_-_38054 | 1.02 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr22_+_110158 | 0.99 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr8_-_48770603 | 0.96 |
ENSDART00000159712
|
pimr184
|
Pim proto-oncogene, serine/threonine kinase, related 184 |
| chr23_-_45319592 | 0.92 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr25_+_16646113 | 0.91 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr13_+_31497236 | 0.86 |
ENSDART00000146752
|
lrrc9
|
leucine rich repeat containing 9 |
| chr14_+_21007957 | 0.86 |
ENSDART00000162316
|
pimr194
|
Pim proto-oncogene, serine/threonine kinase, related 194 |
| chr2_-_879800 | 0.77 |
ENSDART00000019733
|
irf4a
|
interferon regulatory factor 4a |
| chr1_-_30039331 | 0.76 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr19_+_46259619 | 0.75 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr14_-_4121052 | 0.71 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr10_-_27196093 | 0.71 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr14_+_21042178 | 0.69 |
ENSDART00000168551
|
pimr192
|
Pim proto-oncogene, serine/threonine kinase, related 192 |
| chr4_-_149334 | 0.69 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr23_+_31815423 | 0.68 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_-_436658 | 0.67 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr20_+_22045089 | 0.65 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr20_+_48100261 | 0.64 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr16_-_22225295 | 0.64 |
ENSDART00000163519
|
LO017682.1
|
|
| chr8_+_53269657 | 0.61 |
ENSDART00000184212
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr3_-_31619463 | 0.60 |
ENSDART00000124559
|
moto
|
minamoto |
| chr25_-_7723685 | 0.59 |
ENSDART00000146882
|
phf21ab
|
PHD finger protein 21Ab |
| chr23_-_12423778 | 0.59 |
ENSDART00000124091
|
wfdc2
|
WAP four-disulfide core domain 2 |
| chr14_-_31080183 | 0.56 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr25_+_5288665 | 0.56 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr17_+_42027969 | 0.55 |
ENSDART00000147563
|
kiz
|
kizuna centrosomal protein |
| chr3_+_46459540 | 0.54 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr6_-_55254786 | 0.54 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr8_-_7475917 | 0.54 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr20_+_16881883 | 0.53 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr25_+_3347461 | 0.52 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr25_-_13188214 | 0.52 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr9_+_993477 | 0.51 |
ENSDART00000182045
|
CABZ01066921.1
|
|
| chr5_+_13385837 | 0.51 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr23_-_43609595 | 0.50 |
ENSDART00000172222
|
CABZ01117603.1
|
|
| chr9_+_26103814 | 0.50 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr10_+_7029664 | 0.49 |
ENSDART00000166206
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr5_-_65968957 | 0.49 |
ENSDART00000161004
|
ttc16
|
tetratricopeptide repeat domain 16 |
| chr16_-_41004731 | 0.49 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr8_-_38477817 | 0.49 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr14_-_4120636 | 0.49 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr2_+_16780643 | 0.48 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr15_-_25010400 | 0.48 |
ENSDART00000155236
|
si:ch211-198e20.10
|
si:ch211-198e20.10 |
| chr15_-_25083200 | 0.48 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr24_+_41273252 | 0.47 |
ENSDART00000105526
|
nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr1_-_59232267 | 0.47 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr11_-_35763323 | 0.47 |
ENSDART00000026017
|
bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr19_+_7124337 | 0.46 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr6_-_53160459 | 0.46 |
ENSDART00000128127
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr22_+_508290 | 0.45 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr2_+_36004381 | 0.44 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr17_+_14425219 | 0.43 |
ENSDART00000153994
|
ccdc175
|
coiled-coil domain containing 175 |
| chr19_-_7144548 | 0.42 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr8_+_25302172 | 0.42 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr12_+_20641102 | 0.42 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr5_+_37032038 | 0.41 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr11_-_27917730 | 0.40 |
ENSDART00000173219
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr7_+_1383027 | 0.40 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr17_-_51195651 | 0.40 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr22_+_38301365 | 0.40 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr14_+_36889893 | 0.40 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr5_+_28308774 | 0.40 |
ENSDART00000087708
|
si:dkeyp-86b9.1
|
si:dkeyp-86b9.1 |
| chr16_-_24819860 | 0.38 |
ENSDART00000183874
|
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr17_-_10315644 | 0.38 |
ENSDART00000168572
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr5_+_24086227 | 0.38 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr19_+_770300 | 0.37 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr5_-_63425428 | 0.37 |
ENSDART00000163246
|
cntrl
|
centriolin |
| chr18_-_4957022 | 0.36 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr18_-_18543358 | 0.36 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr25_-_1333676 | 0.35 |
ENSDART00000183182
ENSDART00000188907 |
cln6b
|
CLN6, transmembrane ER protein b |
| chr16_-_24832038 | 0.35 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr23_+_19701587 | 0.35 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr11_+_8129536 | 0.34 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr22_-_16180467 | 0.33 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr4_+_2059692 | 0.32 |
ENSDART00000067427
|
yeats4
|
YEATS domain containing 4 |
| chr9_-_889567 | 0.32 |
ENSDART00000155921
|
si:ch73-250a16.5
|
si:ch73-250a16.5 |
| chr22_-_651719 | 0.32 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr12_-_19151708 | 0.32 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr23_-_31974060 | 0.31 |
ENSDART00000168087
|
BX927210.1
|
|
| chr13_-_31452516 | 0.31 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr4_-_12914163 | 0.30 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr16_+_10346277 | 0.30 |
ENSDART00000081092
|
si:dkeyp-77h1.4
|
si:dkeyp-77h1.4 |
| chr19_+_19767567 | 0.30 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr17_-_51893123 | 0.30 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr23_-_32162810 | 0.30 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_-_38861741 | 0.30 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr14_+_50937757 | 0.30 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr1_+_12135129 | 0.29 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr20_-_6196989 | 0.29 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr11_+_35050253 | 0.29 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr14_-_30967284 | 0.28 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr8_+_10823069 | 0.28 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr15_+_27364394 | 0.28 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr25_-_1235457 | 0.27 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr19_-_44801918 | 0.27 |
ENSDART00000108507
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr19_-_7043355 | 0.27 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr17_-_26604946 | 0.27 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr3_-_2591942 | 0.26 |
ENSDART00000127971
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr3_+_35406395 | 0.26 |
ENSDART00000055266
ENSDART00000150918 |
rbbp6
|
retinoblastoma binding protein 6 |
| chr2_-_2642476 | 0.26 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr6_-_442163 | 0.26 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr22_+_28969071 | 0.26 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr3_-_39695856 | 0.26 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr23_-_29003864 | 0.26 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr2_+_2737422 | 0.25 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr21_+_45223194 | 0.25 |
ENSDART00000150902
|
si:ch73-269m14.3
|
si:ch73-269m14.3 |
| chr10_+_31646020 | 0.25 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr9_+_41156287 | 0.24 |
ENSDART00000189195
ENSDART00000186270 |
stat4
|
signal transducer and activator of transcription 4 |
| chr25_+_10458990 | 0.24 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr16_+_13855039 | 0.24 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr8_+_25629722 | 0.24 |
ENSDART00000026807
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr3_+_13190012 | 0.24 |
ENSDART00000179747
ENSDART00000109876 ENSDART00000124824 ENSDART00000130261 |
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr22_+_30184039 | 0.24 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr22_-_34591295 | 0.24 |
ENSDART00000044429
|
rnf123
|
ring finger protein 123 |
| chr23_+_10347851 | 0.23 |
ENSDART00000127667
|
krt18
|
keratin 18 |
| chr25_-_6557854 | 0.23 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr13_+_27316934 | 0.23 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr7_-_28696556 | 0.23 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr4_-_22311610 | 0.23 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr20_-_35076387 | 0.23 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr3_-_2592350 | 0.23 |
ENSDART00000192325
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr21_+_30937690 | 0.22 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr9_+_41156818 | 0.22 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr14_+_49382180 | 0.22 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr15_+_32727848 | 0.22 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr6_-_19333947 | 0.22 |
ENSDART00000160887
|
gga3
|
ggolgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr5_+_30608057 | 0.22 |
ENSDART00000133583
|
si:ch211-117m20.4
|
si:ch211-117m20.4 |
| chr23_-_36418059 | 0.22 |
ENSDART00000135232
|
znf740b
|
zinc finger protein 740b |
| chr13_-_10727550 | 0.22 |
ENSDART00000190925
|
ppm1ba
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ba |
| chr13_-_7474324 | 0.22 |
ENSDART00000162550
|
si:dkey-196h17.9
|
si:dkey-196h17.9 |
| chr7_+_22585447 | 0.21 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr5_+_15819651 | 0.21 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr12_-_18961491 | 0.21 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr7_+_19835569 | 0.21 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr11_+_8565828 | 0.21 |
ENSDART00000126523
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr17_+_23255365 | 0.21 |
ENSDART00000180277
|
AL935174.5
|
|
| chr2_-_5836116 | 0.21 |
ENSDART00000142545
|
mier1b
|
mesoderm induction early response 1b, transcriptional regulator |
| chr5_+_24882633 | 0.21 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr6_-_21616659 | 0.20 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr21_-_22693730 | 0.20 |
ENSDART00000158342
|
gig2d
|
grass carp reovirus (GCRV)-induced gene 2d |
| chr13_+_32148338 | 0.20 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr3_+_35406998 | 0.20 |
ENSDART00000102994
|
rbbp6
|
retinoblastoma binding protein 6 |
| chr10_-_10969444 | 0.20 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr1_+_17892944 | 0.20 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr12_-_30760971 | 0.20 |
ENSDART00000066257
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr15_-_21678634 | 0.20 |
ENSDART00000061139
|
bco2b
|
beta-carotene oxygenase 2b |
| chr25_-_3469576 | 0.19 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr9_+_13733468 | 0.19 |
ENSDART00000165954
ENSDART00000160355 ENSDART00000081040 ENSDART00000138254 ENSDART00000081015 ENSDART00000141314 |
abi2a
|
abl-interactor 2a |
| chr3_-_32170850 | 0.19 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr24_-_33703504 | 0.19 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr2_-_51772438 | 0.19 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr20_+_46371458 | 0.18 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr13_+_30692669 | 0.18 |
ENSDART00000187818
|
CR762483.1
|
|
| chr16_+_23403602 | 0.18 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr24_+_25919809 | 0.18 |
ENSDART00000006615
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr4_-_22519516 | 0.18 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr8_+_22355909 | 0.18 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr14_+_49376011 | 0.18 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr3_-_26524934 | 0.18 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr18_+_27738349 | 0.18 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr2_-_50966124 | 0.18 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr18_+_27821856 | 0.18 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr16_+_50006872 | 0.18 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr14_+_45028062 | 0.18 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr23_+_42819221 | 0.17 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr7_+_56472585 | 0.17 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr6_+_58915889 | 0.17 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr12_+_49100365 | 0.17 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr6_-_35310224 | 0.17 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr6_-_10305918 | 0.17 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr13_-_25819825 | 0.17 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr6_-_49078263 | 0.17 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr16_+_26766423 | 0.17 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr5_-_42883761 | 0.17 |
ENSDART00000167374
|
BX323596.2
|
|
| chr4_+_20263097 | 0.16 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr22_+_3045495 | 0.16 |
ENSDART00000164061
|
LO017843.1
|
|
| chr3_-_40976288 | 0.16 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr5_+_41143563 | 0.16 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr16_+_32995882 | 0.15 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr7_-_40082053 | 0.15 |
ENSDART00000083719
ENSDART00000173687 |
shrprbck1r
|
sharpin and rbck1 related |
| chr16_+_54829293 | 0.15 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr20_-_39367895 | 0.15 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr19_+_19412692 | 0.15 |
ENSDART00000113580
|
wu:fc38h03
|
wu:fc38h03 |
| chr5_+_13373593 | 0.15 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr9_-_52999318 | 0.15 |
ENSDART00000176601
|
CABZ01054965.1
|
|
| chr23_+_4483083 | 0.15 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr7_-_37812176 | 0.14 |
ENSDART00000164485
|
adcy7
|
adenylate cyclase 7 |
| chr10_+_3965565 | 0.14 |
ENSDART00000185405
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr2_+_33052169 | 0.14 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr8_+_53452681 | 0.14 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr2_+_3044992 | 0.14 |
ENSDART00000020463
|
zgc:63882
|
zgc:63882 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfkb1_rela
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.4 | 1.1 | GO:1902230 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.2 | 0.8 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.2 | 1.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.5 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.7 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.4 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.5 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.1 | 0.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 1.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.3 | GO:0034122 | regulation of toll-like receptor signaling pathway(GO:0034121) negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.3 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.1 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.1 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.3 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.1 | 1.3 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.8 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.1 | 0.2 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.6 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 0.7 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.7 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.3 | GO:0035462 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:1903400 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.4 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.1 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.0 | 0.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.0 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.2 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.0 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.1 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.5 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.3 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.4 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.5 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |