Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
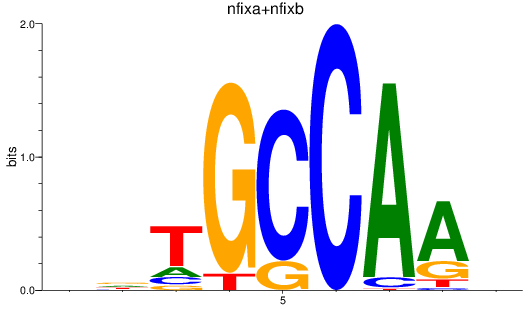
Results for nfixa+nfixb
Z-value: 0.47
Transcription factors associated with nfixa+nfixb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfixa
|
ENSDARG00000043226 | nuclear factor I/Xa |
|
nfixb
|
ENSDARG00000061836 | nuclear factor I/Xb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfixa | dr11_v1_chr1_+_51896147_51896147 | 0.13 | 6.2e-01 | Click! |
| nfixb | dr11_v1_chr3_+_14768364_14768364 | 0.08 | 7.4e-01 | Click! |
Activity profile of nfixa+nfixb motif
Sorted Z-values of nfixa+nfixb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_5207152 | 1.96 |
ENSDART00000172751
|
si:ch73-223f5.2
|
si:ch73-223f5.2 |
| chr25_-_16581233 | 1.80 |
ENSDART00000155312
|
smc1b
|
structural maintenance of chromosomes 1B |
| chr2_-_24488652 | 1.48 |
ENSDART00000052067
|
insl3
|
insulin-like 3 (Leydig cell) |
| chr8_-_52715911 | 1.41 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr13_+_50375800 | 1.40 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr2_-_14798295 | 1.23 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr4_-_8030583 | 1.21 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr7_-_25049377 | 1.15 |
ENSDART00000077191
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr14_+_6954579 | 1.09 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr2_-_24898226 | 1.07 |
ENSDART00000134936
|
si:dkey-149i17.9
|
si:dkey-149i17.9 |
| chr17_-_39786222 | 0.99 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr4_+_17279966 | 0.98 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr12_+_26471712 | 0.89 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr13_+_30804367 | 0.84 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr5_-_51148298 | 0.80 |
ENSDART00000042420
|
ccdc180
|
coiled-coil domain containing 180 |
| chr4_+_72235562 | 0.80 |
ENSDART00000168547
|
si:cabz01071912.2
|
si:cabz01071912.2 |
| chr19_-_25464291 | 0.76 |
ENSDART00000112915
|
umad1
|
UBAP1-MVB12-associated (UMA) domain containing 1 |
| chr6_-_49078263 | 0.69 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr5_+_20693724 | 0.68 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr8_+_46536893 | 0.68 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr17_+_14784275 | 0.67 |
ENSDART00000005355
|
rtraf
|
RNA transcription, translation and transport factor |
| chr22_+_18530395 | 0.67 |
ENSDART00000105415
ENSDART00000183958 |
si:ch211-212d10.1
|
si:ch211-212d10.1 |
| chr16_+_7697878 | 0.67 |
ENSDART00000104176
ENSDART00000172977 |
ccr11.1
|
chemokine (C-C motif) receptor 11.1 |
| chr6_-_9952103 | 0.65 |
ENSDART00000065475
|
zp2l2
|
zona pellucida glycoprotein 2, like 2 |
| chr4_-_8040436 | 0.65 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr18_-_21177674 | 0.64 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr22_-_35330532 | 0.64 |
ENSDART00000172654
|
FO818743.1
|
|
| chr15_+_36966369 | 0.63 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr17_+_33719415 | 0.62 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr18_+_3037998 | 0.61 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr5_+_37087583 | 0.60 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr22_-_15602593 | 0.60 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr2_-_42375275 | 0.58 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr8_+_27555314 | 0.57 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr13_+_15682803 | 0.57 |
ENSDART00000188063
|
CR931980.1
|
|
| chr17_+_21817859 | 0.57 |
ENSDART00000143832
ENSDART00000141462 |
ikzf5
|
IKAROS family zinc finger 5 |
| chr20_+_26880668 | 0.54 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr16_-_13595027 | 0.50 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr11_-_17964525 | 0.50 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr11_-_8262105 | 0.50 |
ENSDART00000173225
|
si:cabz01021066.1
|
si:cabz01021066.1 |
| chr5_-_26181863 | 0.50 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr22_+_34663328 | 0.49 |
ENSDART00000183912
|
si:ch73-304f21.1
|
si:ch73-304f21.1 |
| chr3_-_2592350 | 0.49 |
ENSDART00000192325
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr14_+_39255437 | 0.49 |
ENSDART00000147443
|
diaph2
|
diaphanous-related formin 2 |
| chr21_+_19858627 | 0.48 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr2_-_41861040 | 0.47 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr19_-_7225060 | 0.46 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr16_+_42482270 | 0.45 |
ENSDART00000184225
|
herpud2
|
HERPUD family member 2 |
| chr15_-_39948635 | 0.45 |
ENSDART00000114836
|
msh5
|
mutS homolog 5 |
| chr17_+_35431724 | 0.45 |
ENSDART00000190293
|
BX571665.1
|
|
| chr11_-_22372072 | 0.45 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr3_-_24980067 | 0.45 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr7_-_59054322 | 0.44 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr7_+_59649399 | 0.44 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr22_-_15602760 | 0.43 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr1_-_22851481 | 0.43 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr20_-_47188966 | 0.43 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr3_-_55139127 | 0.43 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr19_-_7144548 | 0.42 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr22_-_37611681 | 0.41 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr3_-_8765165 | 0.40 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr25_-_30429607 | 0.39 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr18_-_48558420 | 0.39 |
ENSDART00000058987
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr5_-_30984271 | 0.39 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr6_+_40661703 | 0.39 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr5_+_57641554 | 0.38 |
ENSDART00000171309
ENSDART00000157992 ENSDART00000164742 |
cryabb
|
crystallin, alpha B, b |
| chr5_-_10239079 | 0.38 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr6_+_32834760 | 0.38 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr20_+_539852 | 0.38 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr16_+_23961276 | 0.37 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr14_+_11457500 | 0.37 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr2_+_38161318 | 0.37 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr8_+_30671060 | 0.36 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr4_+_61171310 | 0.36 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr18_+_44649804 | 0.36 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr24_-_20369760 | 0.34 |
ENSDART00000185585
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr21_-_36619599 | 0.34 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr12_+_26632448 | 0.33 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr11_-_3865472 | 0.33 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr17_-_25563847 | 0.33 |
ENSDART00000040032
|
rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr16_+_23960933 | 0.32 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr24_+_26432541 | 0.32 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr17_+_16873417 | 0.32 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr23_+_6544453 | 0.32 |
ENSDART00000005373
|
spo11
|
SPO11 meiotic protein covalently bound to DSB |
| chr2_-_45510223 | 0.31 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr22_-_11729350 | 0.31 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr14_+_18785727 | 0.31 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr23_-_3674443 | 0.30 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr3_-_39198113 | 0.30 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr7_-_26457208 | 0.29 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr8_-_6877390 | 0.29 |
ENSDART00000170883
ENSDART00000005321 |
neflb
|
neurofilament, light polypeptide b |
| chr13_-_33683889 | 0.28 |
ENSDART00000136820
ENSDART00000065435 |
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr14_+_28438947 | 0.28 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr25_-_18002937 | 0.28 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr4_-_38033800 | 0.27 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr7_+_25049545 | 0.26 |
ENSDART00000173896
ENSDART00000173566 |
si:dkey-23i12.7
ppp2r5b
|
si:dkey-23i12.7 protein phosphatase 2, regulatory subunit B', beta |
| chr10_+_5025696 | 0.26 |
ENSDART00000031165
|
enoph1
|
enolase-phosphatase 1 |
| chr18_-_17529763 | 0.26 |
ENSDART00000191308
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr9_-_2572790 | 0.25 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr8_-_20138054 | 0.25 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr21_+_27448856 | 0.25 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr17_-_8886735 | 0.25 |
ENSDART00000121997
|
nkl.3
|
NK-lysin tandem duplicate 3 |
| chr4_-_12388535 | 0.25 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr24_-_38574631 | 0.24 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr9_+_13999620 | 0.24 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr15_-_28082310 | 0.24 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr14_+_11458044 | 0.23 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr7_+_35238234 | 0.23 |
ENSDART00000178732
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr21_-_25555355 | 0.23 |
ENSDART00000144228
|
si:dkey-17e16.9
|
si:dkey-17e16.9 |
| chr8_+_48858132 | 0.22 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr25_-_30394746 | 0.22 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr14_-_21064199 | 0.22 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr1_+_22851261 | 0.22 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr7_+_57108823 | 0.22 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr18_-_48983690 | 0.22 |
ENSDART00000182359
|
FO681288.3
|
|
| chr9_-_22963897 | 0.21 |
ENSDART00000133676
|
si:dkey-91i10.2
|
si:dkey-91i10.2 |
| chr25_+_27923846 | 0.21 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr22_+_19553390 | 0.21 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr14_+_25465346 | 0.21 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr4_+_70556298 | 0.21 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr20_+_32497260 | 0.21 |
ENSDART00000145175
ENSDART00000132921 |
si:ch73-257c13.2
|
si:ch73-257c13.2 |
| chr20_+_38525567 | 0.21 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
| chr19_-_24163845 | 0.20 |
ENSDART00000133277
|
prf1.2
|
perforin 1.2 |
| chr12_-_6880694 | 0.20 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr3_-_26921475 | 0.20 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr13_-_20381485 | 0.20 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr9_+_51147764 | 0.20 |
ENSDART00000188480
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr16_-_28876479 | 0.19 |
ENSDART00000047862
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr18_+_17660158 | 0.19 |
ENSDART00000186279
|
cpne2
|
copine II |
| chr19_+_30387999 | 0.19 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr10_+_39263827 | 0.19 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr11_-_1933793 | 0.19 |
ENSDART00000186850
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr6_-_35052145 | 0.19 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr1_-_51684941 | 0.19 |
ENSDART00000113853
|
prokr1a
|
prokineticin receptor 1a |
| chr6_-_24301324 | 0.18 |
ENSDART00000171401
|
BX927081.1
|
|
| chr19_-_12404590 | 0.18 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr15_-_18361475 | 0.18 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr16_-_22713152 | 0.17 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr16_+_46430627 | 0.17 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr9_+_54237100 | 0.17 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr1_-_46862190 | 0.17 |
ENSDART00000145167
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr13_+_17672527 | 0.17 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr22_+_5478353 | 0.17 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr21_-_4032650 | 0.17 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr21_-_2958422 | 0.16 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr6_-_8580857 | 0.16 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr17_-_48743076 | 0.15 |
ENSDART00000173117
|
kcnk17
|
potassium channel, subfamily K, member 17 |
| chr24_-_28381404 | 0.15 |
ENSDART00000148406
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr19_-_29832876 | 0.14 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr2_-_14793343 | 0.14 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr3_+_54744069 | 0.14 |
ENSDART00000134958
ENSDART00000114443 |
si:ch211-74m13.3
|
si:ch211-74m13.3 |
| chr1_-_54706039 | 0.14 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr17_-_14451405 | 0.14 |
ENSDART00000146786
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr16_-_49846359 | 0.14 |
ENSDART00000184246
|
znf385d
|
zinc finger protein 385D |
| chr2_-_58193381 | 0.14 |
ENSDART00000159040
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr19_+_30990815 | 0.14 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr5_-_57641257 | 0.13 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr23_+_13721826 | 0.13 |
ENSDART00000142494
|
zbtb46
|
zinc finger and BTB domain containing 46 |
| chr1_-_58900851 | 0.13 |
ENSDART00000183085
ENSDART00000188855 ENSDART00000182567 |
CABZ01084501.3
|
Danio rerio microfibril-associated glycoprotein 4-like (LOC100334800), transcript variant 2, mRNA. |
| chr11_+_44579865 | 0.13 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr22_+_5176255 | 0.13 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr8_+_52642869 | 0.12 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr9_+_6587364 | 0.12 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr16_-_29458806 | 0.12 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr9_-_39968820 | 0.12 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr3_-_32603191 | 0.12 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr5_+_45918680 | 0.12 |
ENSDART00000036242
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr18_+_17660402 | 0.12 |
ENSDART00000143475
|
cpne2
|
copine II |
| chr15_-_40246396 | 0.11 |
ENSDART00000063777
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr22_-_28653074 | 0.11 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr7_-_21928826 | 0.11 |
ENSDART00000088043
|
si:dkey-85k7.11
|
si:dkey-85k7.11 |
| chr6_+_39905021 | 0.11 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr8_+_28469054 | 0.11 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr1_-_9644630 | 0.11 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr8_+_31016180 | 0.11 |
ENSDART00000130870
ENSDART00000143604 |
odf2b
|
outer dense fiber of sperm tails 2b |
| chr12_-_37941733 | 0.11 |
ENSDART00000130167
|
CABZ01046949.1
|
|
| chr22_+_18816662 | 0.11 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr22_-_13544244 | 0.10 |
ENSDART00000110136
|
cntnap5b
|
contactin associated protein-like 5b |
| chr22_+_12477996 | 0.10 |
ENSDART00000177704
|
CR847870.3
|
|
| chr23_+_44741500 | 0.10 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr11_-_13151841 | 0.10 |
ENSDART00000161983
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr25_-_19068557 | 0.10 |
ENSDART00000184780
|
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr2_-_37889111 | 0.10 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr19_+_10603405 | 0.10 |
ENSDART00000151135
|
si:dkey-211g8.8
|
si:dkey-211g8.8 |
| chr22_+_5176693 | 0.10 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr19_-_24443867 | 0.10 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr6_-_21873266 | 0.10 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr20_-_29229843 | 0.10 |
ENSDART00000101418
ENSDART00000184637 |
chrm5b
|
cholinergic receptor, muscarinic 5b |
| chr10_-_26738209 | 0.10 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr19_+_4463389 | 0.09 |
ENSDART00000168805
|
kcnk9
|
potassium channel, subfamily K, member 9 |
| chr16_+_48682309 | 0.09 |
ENSDART00000183957
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr13_+_18545819 | 0.09 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr19_+_10831362 | 0.09 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr4_+_29206813 | 0.09 |
ENSDART00000131893
|
si:dkey-23a23.1
|
si:dkey-23a23.1 |
| chr10_-_36231741 | 0.09 |
ENSDART00000170641
|
or109-4
|
odorant receptor, family D, subfamily 109, member 4 |
| chr15_-_29387446 | 0.09 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr21_-_18993110 | 0.09 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr3_+_52467879 | 0.09 |
ENSDART00000156039
|
adgre5a
|
adhesion G protein-coupled receptor E5a |
| chr22_+_11857356 | 0.09 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr21_+_15866522 | 0.08 |
ENSDART00000110728
|
gcnt4b
|
glucosaminyl (N-acetyl) transferase 4, core 2, b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfixa+nfixb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.3 | 0.8 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.2 | 1.0 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 0.7 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 1.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.4 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.5 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.3 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.5 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) pronephric duct development(GO:0039022) |
| 0.0 | 0.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 1.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 2.8 | GO:0070646 | protein modification by small protein removal(GO:0070646) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:1903170 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of voltage-gated calcium channel activity(GO:1901386) negative regulation of calcium ion transmembrane transport(GO:1903170) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 1.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.8 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 1.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0046977 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.1 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 2.0 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |