Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
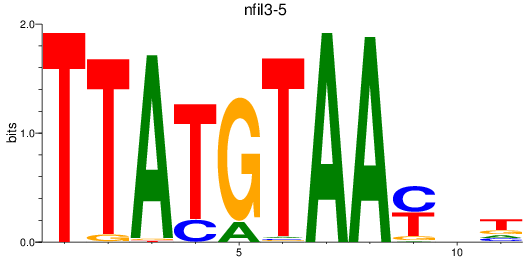
Results for nfil3-5
Z-value: 0.19
Transcription factors associated with nfil3-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfil3-5
|
ENSDARG00000094965 | nuclear factor, interleukin 3 regulated, member 5 |
|
nfil3-5
|
ENSDARG00000113345 | nuclear factor, interleukin 3 regulated, member 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfil3-5 | dr11_v1_chr6_+_8172227_8172227 | 0.11 | 6.7e-01 | Click! |
Activity profile of nfil3-5 motif
Sorted Z-values of nfil3-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_22974019 | 0.19 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr5_-_65153731 | 0.17 |
ENSDART00000171024
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr18_-_39288894 | 0.16 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr9_-_48736388 | 0.13 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr2_+_16597011 | 0.11 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr6_-_21616659 | 0.11 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr22_+_10606863 | 0.11 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr22_+_786556 | 0.11 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr13_-_33683889 | 0.11 |
ENSDART00000136820
ENSDART00000065435 |
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr22_+_10606573 | 0.11 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr13_+_7049823 | 0.11 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr24_-_17023392 | 0.11 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr17_-_8268406 | 0.10 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr16_-_33105677 | 0.10 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr1_+_16600690 | 0.10 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr11_-_33612433 | 0.10 |
ENSDART00000179756
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr23_+_33963619 | 0.10 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr21_+_43178831 | 0.10 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr5_-_69716501 | 0.09 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr7_-_40959667 | 0.09 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr10_+_26944418 | 0.09 |
ENSDART00000135493
|
frmd8
|
FERM domain containing 8 |
| chr7_+_38380135 | 0.08 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr14_+_6962271 | 0.08 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr8_-_21103522 | 0.08 |
ENSDART00000100283
|
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr4_+_5333988 | 0.08 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr11_+_41090440 | 0.08 |
ENSDART00000188287
|
phf13
|
PHD finger protein 13 |
| chr8_+_47897734 | 0.08 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr3_-_26184018 | 0.08 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_55848358 | 0.08 |
ENSDART00000130891
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr3_-_26183699 | 0.08 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr2_-_2642476 | 0.08 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr21_-_32467799 | 0.08 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr5_-_55848511 | 0.08 |
ENSDART00000183503
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr22_+_15973122 | 0.07 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr23_-_32194397 | 0.07 |
ENSDART00000184206
ENSDART00000166682 |
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr14_-_21618005 | 0.07 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr13_+_7049210 | 0.07 |
ENSDART00000168608
ENSDART00000128675 ENSDART00000164789 |
rnaset2
|
ribonuclease T2 |
| chr4_+_5334439 | 0.07 |
ENSDART00000180644
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr13_-_36798204 | 0.07 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr23_-_21763598 | 0.07 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr20_-_36617313 | 0.07 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr15_+_21882419 | 0.07 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr15_+_24737599 | 0.07 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr4_+_3389866 | 0.07 |
ENSDART00000153834
|
sypl1
|
synaptophysin-like 1 |
| chr19_+_7549854 | 0.07 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr10_+_18911152 | 0.07 |
ENSDART00000030205
|
bnip3lb
|
BCL2 interacting protein 3 like b |
| chr4_+_11690923 | 0.07 |
ENSDART00000150624
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr8_-_21103041 | 0.07 |
ENSDART00000171771
ENSDART00000131322 ENSDART00000137838 |
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr9_-_34509997 | 0.07 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr8_+_21225064 | 0.07 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr2_+_2168547 | 0.07 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr21_-_32467099 | 0.07 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr21_+_11923701 | 0.07 |
ENSDART00000109292
|
ubap2a
|
ubiquitin associated protein 2a |
| chr6_+_16468776 | 0.07 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr23_-_21534455 | 0.07 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr16_-_27676123 | 0.07 |
ENSDART00000180804
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr25_-_12803723 | 0.07 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr23_-_21534738 | 0.06 |
ENSDART00000134587
|
rcc2
|
regulator of chromosome condensation 2 |
| chr23_-_33361425 | 0.06 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr8_-_11324143 | 0.06 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr4_-_27099224 | 0.06 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr23_-_21535040 | 0.06 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr5_+_43782267 | 0.06 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr13_-_27354003 | 0.06 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr20_-_52939501 | 0.06 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr16_-_31790285 | 0.06 |
ENSDART00000184655
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr7_-_48396193 | 0.06 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr9_+_45227028 | 0.06 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr21_-_26677834 | 0.06 |
ENSDART00000077381
|
nxf1
|
nuclear RNA export factor 1 |
| chr6_-_13022166 | 0.06 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr19_+_23982466 | 0.06 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr5_-_14521500 | 0.06 |
ENSDART00000176565
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr10_-_39283883 | 0.06 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr7_-_38861741 | 0.06 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr7_+_38278860 | 0.06 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr16_-_19568388 | 0.06 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr18_-_14879135 | 0.06 |
ENSDART00000099701
|
selenoo1
|
selenoprotein O1 |
| chr22_-_13165186 | 0.06 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr3_+_7763114 | 0.05 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr20_+_38671894 | 0.05 |
ENSDART00000146544
|
mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr22_-_4439311 | 0.05 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr6_-_32411703 | 0.05 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr20_+_25711718 | 0.05 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr15_-_4969525 | 0.05 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr3_+_33345348 | 0.05 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr17_+_25833947 | 0.05 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr16_+_23397785 | 0.05 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr14_+_16034447 | 0.05 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr20_+_47953047 | 0.05 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr20_+_1121458 | 0.05 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr19_-_46037835 | 0.05 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr9_-_34500197 | 0.05 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr3_-_5030089 | 0.05 |
ENSDART00000167153
|
polr3h
|
polymerase (RNA) III (DNA directed) polypeptide H |
| chr1_-_45616242 | 0.05 |
ENSDART00000150066
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr11_+_24348425 | 0.05 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr15_-_31419805 | 0.05 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr17_-_5610514 | 0.05 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr19_-_46018152 | 0.05 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr4_+_5334202 | 0.05 |
ENSDART00000150409
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr16_-_35427060 | 0.05 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr15_+_28303161 | 0.05 |
ENSDART00000087926
|
myo1cb
|
myosin Ic, paralog b |
| chr3_+_22335030 | 0.05 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr23_+_20640875 | 0.05 |
ENSDART00000147382
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr20_+_38201644 | 0.05 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr25_-_28668776 | 0.05 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr7_+_17816006 | 0.05 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr16_-_41646164 | 0.05 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr19_+_8612839 | 0.05 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr22_-_3595439 | 0.05 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr12_-_13650344 | 0.05 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr3_-_5228841 | 0.05 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr21_-_2261720 | 0.05 |
ENSDART00000170161
|
si:ch73-299h12.2
|
si:ch73-299h12.2 |
| chr4_+_9177997 | 0.05 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr16_-_21047872 | 0.04 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr7_-_38570299 | 0.04 |
ENSDART00000143815
|
celf1
|
cugbp, Elav-like family member 1 |
| chr1_+_19764995 | 0.04 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr18_-_38271298 | 0.04 |
ENSDART00000143016
|
caprin1b
|
cell cycle associated protein 1b |
| chr10_+_39283985 | 0.04 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr14_+_23717165 | 0.04 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr25_+_33192796 | 0.04 |
ENSDART00000125892
ENSDART00000121680 ENSDART00000014851 |
TPM1 (1 of many)
|
zgc:171719 |
| chr11_-_35171768 | 0.04 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr7_-_69429561 | 0.04 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr3_-_15144067 | 0.04 |
ENSDART00000127738
ENSDART00000060426 ENSDART00000180799 |
fam173a
|
family with sequence similarity 173, member A |
| chr23_-_18981595 | 0.04 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr19_+_5674907 | 0.04 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr6_-_7726849 | 0.04 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr14_+_21699414 | 0.04 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr24_-_39772045 | 0.04 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr16_+_23398369 | 0.04 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr9_+_27720428 | 0.04 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr16_+_40954481 | 0.04 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr6_-_1187749 | 0.04 |
ENSDART00000172544
|
txnrd3
|
thioredoxin reductase 3 |
| chr3_+_1107102 | 0.04 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr18_+_7456597 | 0.04 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr2_+_27830436 | 0.04 |
ENSDART00000182253
|
FO834800.2
|
|
| chr24_+_37688729 | 0.04 |
ENSDART00000137017
|
h3f3d
|
H3 histone, family 3D |
| chr4_+_5334707 | 0.04 |
ENSDART00000150620
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr9_+_34433053 | 0.04 |
ENSDART00000137416
|
si:ch211-269e2.1
|
si:ch211-269e2.1 |
| chr8_+_13700605 | 0.04 |
ENSDART00000144516
|
lonrf1l
|
LON peptidase N-terminal domain and ring finger 1, like |
| chr13_+_30035253 | 0.04 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr16_-_33104944 | 0.04 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr25_-_25575717 | 0.04 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr6_-_12314475 | 0.04 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr9_+_46644633 | 0.04 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr6_-_1187565 | 0.04 |
ENSDART00000191756
|
txnrd3
|
thioredoxin reductase 3 |
| chr9_+_29520696 | 0.04 |
ENSDART00000144430
|
fdx1
|
ferredoxin 1 |
| chr5_-_30074332 | 0.04 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr6_-_39521832 | 0.04 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr14_+_21699129 | 0.03 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr22_-_10605045 | 0.03 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr20_-_23876291 | 0.03 |
ENSDART00000043316
|
katna1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr17_-_23709347 | 0.03 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr23_+_13528053 | 0.03 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr6_-_39649504 | 0.03 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr21_+_5960443 | 0.03 |
ENSDART00000149689
|
mob1bb
|
MOB kinase activator 1Bb |
| chr18_+_41561285 | 0.03 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr11_-_37720024 | 0.03 |
ENSDART00000023388
|
yod1
|
YOD1 deubiquitinase |
| chr11_+_714386 | 0.03 |
ENSDART00000167824
ENSDART00000187110 |
timp4.3
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 3 |
| chr25_+_33192404 | 0.03 |
ENSDART00000193592
|
TPM1 (1 of many)
|
zgc:171719 |
| chr25_-_2723682 | 0.03 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr24_+_23840821 | 0.03 |
ENSDART00000128595
ENSDART00000127188 |
pola1
|
polymerase (DNA directed), alpha 1 |
| chr5_-_38248347 | 0.03 |
ENSDART00000084917
ENSDART00000139479 |
slc12a9
|
solute carrier family 12, member 9 |
| chr18_+_6536598 | 0.03 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr23_+_13528550 | 0.03 |
ENSDART00000099903
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr20_-_30900947 | 0.03 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr16_-_12097558 | 0.03 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr8_+_17869225 | 0.03 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr6_-_23002373 | 0.03 |
ENSDART00000037709
ENSDART00000170369 |
nol11
|
nucleolar protein 11 |
| chr16_-_12097394 | 0.03 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr24_-_23784701 | 0.03 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr19_+_4888491 | 0.03 |
ENSDART00000151540
|
cdk12
|
cyclin-dependent kinase 12 |
| chr7_+_13639473 | 0.03 |
ENSDART00000157870
ENSDART00000032899 |
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr6_+_3280939 | 0.03 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr6_+_9793495 | 0.03 |
ENSDART00000108524
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr4_-_14915268 | 0.03 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr7_-_32782430 | 0.03 |
ENSDART00000173808
|
gas2b
|
growth arrest-specific 2b |
| chr7_+_34297271 | 0.03 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_18920213 | 0.03 |
ENSDART00000193777
|
rnf38
|
ring finger protein 38 |
| chr4_-_11024767 | 0.03 |
ENSDART00000067261
ENSDART00000167631 |
tmtc2a
|
transmembrane and tetratricopeptide repeat containing 2a |
| chr1_+_11881559 | 0.03 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr2_+_5843694 | 0.03 |
ENSDART00000182364
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr21_+_4388489 | 0.03 |
ENSDART00000144555
|
HTRA2 (1 of many)
|
si:dkey-84o3.3 |
| chr3_+_28594713 | 0.03 |
ENSDART00000179799
|
sept12
|
septin 12 |
| chr18_+_26899316 | 0.03 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr16_+_13883872 | 0.03 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr19_-_12145390 | 0.03 |
ENSDART00000143087
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr16_+_10841163 | 0.03 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr19_-_12145765 | 0.03 |
ENSDART00000032474
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr16_+_34493987 | 0.03 |
ENSDART00000138374
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr16_+_25296389 | 0.03 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr12_+_8074343 | 0.03 |
ENSDART00000124084
|
cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr2_+_16696052 | 0.03 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr11_-_7078392 | 0.03 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr15_+_7992906 | 0.03 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr8_+_7801060 | 0.03 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr25_+_34014523 | 0.03 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr23_+_37323962 | 0.03 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr5_+_62340799 | 0.03 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr25_-_10630496 | 0.03 |
ENSDART00000153639
ENSDART00000181722 ENSDART00000177834 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr15_-_19128705 | 0.02 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfil3-5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0070197 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.1 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 0.0 | GO:0045601 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.0 | 0.0 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.0 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.0 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.0 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0030826 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.0 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.0 | GO:0097268 | cytoophidium(GO:0097268) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.1 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |