Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
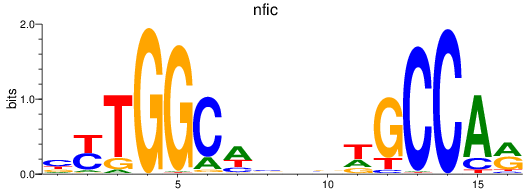
Results for nfic
Z-value: 0.09
Transcription factors associated with nfic
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfic
|
ENSDARG00000043210 | nuclear factor I/C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfic | dr11_v1_chr8_+_20679759_20679759 | -0.62 | 6.0e-03 | Click! |
Activity profile of nfic motif
Sorted Z-values of nfic motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_19467011 | 0.17 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr2_-_21167652 | 0.16 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr25_-_29074064 | 0.14 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr16_-_48428035 | 0.14 |
ENSDART00000190554
|
rad21a
|
RAD21 cohesin complex component a |
| chr20_+_48739154 | 0.13 |
ENSDART00000100262
|
zgc:110348
|
zgc:110348 |
| chr8_-_25033681 | 0.12 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr25_-_36261836 | 0.12 |
ENSDART00000179411
|
dus2
|
dihydrouridine synthase 2 |
| chr19_-_33996277 | 0.12 |
ENSDART00000159384
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr3_-_43646733 | 0.11 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr6_-_6254432 | 0.11 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr12_-_35386910 | 0.11 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr21_+_38089036 | 0.10 |
ENSDART00000147219
|
klf8
|
Kruppel-like factor 8 |
| chr11_-_24538852 | 0.10 |
ENSDART00000171004
ENSDART00000181039 |
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr23_-_26227805 | 0.09 |
ENSDART00000158082
|
BX927204.1
|
|
| chr2_-_40889465 | 0.09 |
ENSDART00000192631
ENSDART00000180824 |
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr14_+_21820034 | 0.08 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr8_+_9699111 | 0.08 |
ENSDART00000111853
|
gripap1
|
GRIP1 associated protein 1 |
| chr15_-_20745513 | 0.08 |
ENSDART00000187438
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr6_+_19933763 | 0.08 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr8_+_44623540 | 0.07 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr19_+_7636941 | 0.07 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr19_-_5332784 | 0.07 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr10_-_40939706 | 0.07 |
ENSDART00000059795
ENSDART00000190510 |
bmp1b
|
bone morphogenetic protein 1b |
| chr1_+_35194454 | 0.07 |
ENSDART00000165389
|
sc:d189
|
sc:d189 |
| chr13_+_18371208 | 0.06 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr8_-_44223899 | 0.06 |
ENSDART00000143020
|
stx2b
|
syntaxin 2b |
| chr10_-_40939303 | 0.06 |
ENSDART00000134295
|
bmp1b
|
bone morphogenetic protein 1b |
| chr7_+_31051603 | 0.06 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr16_+_33143503 | 0.06 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr8_-_44223473 | 0.06 |
ENSDART00000098525
|
stx2b
|
syntaxin 2b |
| chr8_-_1266181 | 0.06 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr13_+_22731356 | 0.06 |
ENSDART00000133064
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr20_-_34069956 | 0.06 |
ENSDART00000017941
|
tprb
|
translocated promoter region b, nuclear basket protein |
| chr16_+_21426524 | 0.06 |
ENSDART00000182869
|
gsdmeb
|
gasdermin Eb |
| chr15_+_24749985 | 0.06 |
ENSDART00000146107
|
ywhae1
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 |
| chr4_+_12292274 | 0.06 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr19_+_14109348 | 0.06 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr23_-_20175674 | 0.06 |
ENSDART00000112840
ENSDART00000189562 |
usp19
|
ubiquitin specific peptidase 19 |
| chr20_+_9474841 | 0.05 |
ENSDART00000053847
|
rad51b
|
RAD51 paralog B |
| chr20_+_54333774 | 0.05 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr9_-_41323746 | 0.05 |
ENSDART00000140564
|
glsb
|
glutaminase b |
| chr6_+_7533601 | 0.05 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr20_-_3390406 | 0.05 |
ENSDART00000136987
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr7_+_38962459 | 0.05 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr1_+_28089703 | 0.05 |
ENSDART00000166989
|
psip1a
|
PC4 and SFRS1 interacting protein 1a |
| chr5_-_37044262 | 0.05 |
ENSDART00000166670
|
si:dkeyp-110c7.8
|
si:dkeyp-110c7.8 |
| chr16_+_35401543 | 0.05 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr5_-_54772982 | 0.04 |
ENSDART00000122829
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr11_-_17755444 | 0.04 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_-_38777553 | 0.04 |
ENSDART00000193045
|
znf281a
|
zinc finger protein 281a |
| chr7_-_28647959 | 0.04 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr7_+_36898850 | 0.04 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr3_-_15667713 | 0.04 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr12_+_48511605 | 0.04 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr22_-_29083070 | 0.04 |
ENSDART00000104812
ENSDART00000172576 |
cbx6a
|
chromobox homolog 6a |
| chr18_-_42313798 | 0.04 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr16_+_30604387 | 0.04 |
ENSDART00000058785
|
fam210ab
|
family with sequence similarity 210, member Ab |
| chr8_-_25566347 | 0.04 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr3_-_33934788 | 0.03 |
ENSDART00000151411
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr5_-_32338866 | 0.03 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_30725473 | 0.03 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr19_+_40379771 | 0.03 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr25_+_2993855 | 0.03 |
ENSDART00000163009
|
neo1b
|
neogenin 1b |
| chr8_-_31053872 | 0.03 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr21_-_22681534 | 0.03 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr8_-_16515127 | 0.03 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr10_-_29827000 | 0.03 |
ENSDART00000131418
|
zpr1
|
ZPR1 zinc finger |
| chr2_-_38114370 | 0.03 |
ENSDART00000131837
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr9_-_32142311 | 0.03 |
ENSDART00000142768
|
ankrd44
|
ankyrin repeat domain 44 |
| chr12_+_30653047 | 0.03 |
ENSDART00000148562
|
thbs2b
|
thrombospondin 2b |
| chr7_+_36898622 | 0.03 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr3_-_37681824 | 0.03 |
ENSDART00000185858
|
gpatch8
|
G patch domain containing 8 |
| chr14_-_25444774 | 0.03 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr18_+_9493720 | 0.03 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr3_-_61385117 | 0.02 |
ENSDART00000170164
|
znf1028
|
zinc finger protein 1028 |
| chr10_-_1718395 | 0.02 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr6_-_39199070 | 0.02 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr5_+_68826514 | 0.02 |
ENSDART00000061406
|
usp39
|
ubiquitin specific peptidase 39 |
| chr6_-_35032792 | 0.02 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr21_+_10866421 | 0.02 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr7_+_37377335 | 0.02 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr23_+_21978584 | 0.02 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr24_-_35707552 | 0.02 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_+_28683574 | 0.02 |
ENSDART00000152000
|
hectd1
|
HECT domain containing 1 |
| chr17_-_50050453 | 0.02 |
ENSDART00000182057
|
zgc:100951
|
zgc:100951 |
| chr5_+_37517800 | 0.02 |
ENSDART00000048107
|
FP102018.1
|
Danio rerio latent transforming growth factor beta binding protein 3 (ltbp3), mRNA. |
| chr4_+_30718997 | 0.02 |
ENSDART00000190901
|
CU207245.1
|
|
| chr25_+_4660639 | 0.02 |
ENSDART00000130299
|
deaf1
|
DEAF1 transcription factor |
| chr10_+_36345176 | 0.02 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr3_-_40275096 | 0.02 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr6_+_56076595 | 0.02 |
ENSDART00000169747
|
rtf2
|
replication termination factor 2 domain containing 1 |
| chr14_+_30775066 | 0.02 |
ENSDART00000139975
ENSDART00000144921 |
atl3
|
atlastin 3 |
| chr20_+_23960525 | 0.01 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr5_+_23211938 | 0.01 |
ENSDART00000090102
|
tpcn1
|
two pore segment channel 1 |
| chr5_-_67629263 | 0.01 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr19_-_9786914 | 0.01 |
ENSDART00000181669
|
si:dkey-14o18.2
|
si:dkey-14o18.2 |
| chr16_-_43041324 | 0.01 |
ENSDART00000155445
ENSDART00000156836 ENSDART00000154945 |
si:dkey-7j14.6
|
si:dkey-7j14.6 |
| chr8_-_36399884 | 0.01 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr17_-_35352690 | 0.01 |
ENSDART00000016053
|
rnf144aa
|
ring finger protein 144aa |
| chr22_-_15693085 | 0.01 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr13_-_22774491 | 0.01 |
ENSDART00000189320
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr20_-_25486384 | 0.01 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr19_+_9786722 | 0.01 |
ENSDART00000138310
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr1_+_4101741 | 0.01 |
ENSDART00000163793
|
slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr10_+_40684758 | 0.01 |
ENSDART00000133648
|
taar19h
|
trace amine associated receptor 19h |
| chr2_+_52049239 | 0.01 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr7_+_52135791 | 0.01 |
ENSDART00000098705
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr23_-_20363261 | 0.01 |
ENSDART00000054651
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr15_-_31508221 | 0.01 |
ENSDART00000121464
|
si:dkey-1m11.5
|
si:dkey-1m11.5 |
| chr4_+_74131530 | 0.01 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr1_-_28089557 | 0.01 |
ENSDART00000161024
ENSDART00000167875 |
snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr13_+_33651416 | 0.01 |
ENSDART00000180221
|
BX005372.1
|
|
| chr21_-_34658266 | 0.01 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr17_+_5846202 | 0.01 |
ENSDART00000189713
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr2_-_25159309 | 0.01 |
ENSDART00000137290
|
si:dkey-223d7.6
|
si:dkey-223d7.6 |
| chr5_-_37935607 | 0.01 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr11_-_43948885 | 0.01 |
ENSDART00000164522
|
CABZ01074203.1
|
|
| chr5_+_55225497 | 0.01 |
ENSDART00000144087
|
tmc2a
|
transmembrane channel-like 2a |
| chr21_-_22678195 | 0.01 |
ENSDART00000171231
|
gig2g
|
grass carp reovirus (GCRV)-induced gene 2g |
| chr21_+_43253538 | 0.01 |
ENSDART00000179940
ENSDART00000164806 ENSDART00000147026 |
shroom1
|
shroom family member 1 |
| chr13_+_40635844 | 0.01 |
ENSDART00000137310
|
hpse2
|
heparanase 2 |
| chr8_-_14375890 | 0.01 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr15_-_18091157 | 0.01 |
ENSDART00000153848
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr20_-_8110672 | 0.01 |
ENSDART00000113993
|
si:ch211-232i5.1
|
si:ch211-232i5.1 |
| chr22_+_2623006 | 0.01 |
ENSDART00000176556
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr7_+_27898091 | 0.01 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr7_+_52122224 | 0.01 |
ENSDART00000174268
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr22_+_2143107 | 0.01 |
ENSDART00000172339
ENSDART00000161280 ENSDART00000163671 ENSDART00000172090 ENSDART00000163573 |
znf1164
|
zinc finger protein 1164 |
| chr21_+_11865972 | 0.00 |
ENSDART00000081676
|
ubap1
|
ubiquitin associated protein 1 |
| chr3_+_22905341 | 0.00 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr11_+_14284866 | 0.00 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr20_-_26588736 | 0.00 |
ENSDART00000134337
|
exoc2
|
exocyst complex component 2 |
| chr22_-_7129631 | 0.00 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr22_+_2144278 | 0.00 |
ENSDART00000162173
ENSDART00000159914 ENSDART00000160192 |
znf1164
|
zinc finger protein 1164 |
| chr15_-_18200358 | 0.00 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr4_+_1283068 | 0.00 |
ENSDART00000167233
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr18_+_33818577 | 0.00 |
ENSDART00000131788
|
olfcq19
|
olfactory receptor C family, q19 |
| chr7_+_9008372 | 0.00 |
ENSDART00000144897
|
AL935128.1
|
|
| chr8_+_43852743 | 0.00 |
ENSDART00000186485
|
AL808129.2
|
|
| chr3_+_50602866 | 0.00 |
ENSDART00000190069
ENSDART00000153921 |
gsg1l2a
|
gsg1-like 2a |
| chr10_-_43844537 | 0.00 |
ENSDART00000114208
|
tlr8b
|
toll-like receptor 8b |
| chr4_-_75899294 | 0.00 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr9_+_12934536 | 0.00 |
ENSDART00000134484
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr20_+_51464670 | 0.00 |
ENSDART00000150110
|
thbd
|
thrombomodulin |
| chr9_-_46382637 | 0.00 |
ENSDART00000085738
|
lct
|
lactase |
| chr4_+_59589201 | 0.00 |
ENSDART00000150576
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
| chr25_-_17367578 | 0.00 |
ENSDART00000064591
ENSDART00000110692 |
cyp2x6
|
cytochrome P450, family 2, subfamily X, polypeptide 6 |
| chr7_+_73447724 | 0.00 |
ENSDART00000040786
|
cbln6
|
cerebellin 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfic
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |