Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
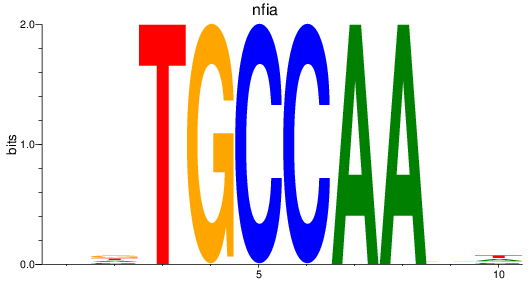
Results for nfia
Z-value: 0.50
Transcription factors associated with nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfia
|
ENSDARG00000062420 | nuclear factor I/A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfia | dr11_v1_chr22_-_16997475_16997475 | 0.59 | 1.0e-02 | Click! |
Activity profile of nfia motif
Sorted Z-values of nfia motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_50375800 | 1.23 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr3_+_31039923 | 1.15 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr13_+_30804367 | 0.91 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr25_-_10503043 | 0.91 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr6_-_49078263 | 0.81 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr1_-_58900851 | 0.77 |
ENSDART00000183085
ENSDART00000188855 ENSDART00000182567 |
CABZ01084501.3
|
Danio rerio microfibril-associated glycoprotein 4-like (LOC100334800), transcript variant 2, mRNA. |
| chr3_+_55105066 | 0.77 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr3_+_55100045 | 0.77 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr24_-_38197040 | 0.76 |
ENSDART00000137949
ENSDART00000105639 |
igic1s1
|
immunoglobulin light iota constant 1, s1 |
| chr14_-_3268155 | 0.74 |
ENSDART00000177244
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr8_-_52715911 | 0.72 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr17_-_39786222 | 0.72 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr25_-_16581233 | 0.71 |
ENSDART00000155312
|
smc1b
|
structural maintenance of chromosomes 1B |
| chr20_+_25340814 | 0.70 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr3_-_30962646 | 0.70 |
ENSDART00000110861
|
paqr4a
|
progestin and adipoQ receptor family member IVa |
| chr2_-_14798295 | 0.65 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr7_-_5207152 | 0.63 |
ENSDART00000172751
|
si:ch73-223f5.2
|
si:ch73-223f5.2 |
| chr16_+_7697878 | 0.62 |
ENSDART00000104176
ENSDART00000172977 |
ccr11.1
|
chemokine (C-C motif) receptor 11.1 |
| chr6_-_21091948 | 0.62 |
ENSDART00000057348
|
inha
|
inhibin, alpha |
| chr16_-_13595027 | 0.62 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr6_+_269204 | 0.61 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr15_-_34567370 | 0.61 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr5_+_42467867 | 0.59 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr24_-_5910811 | 0.58 |
ENSDART00000163012
|
pcolce2b
|
procollagen C-endopeptidase enhancer 2b |
| chr21_-_20932603 | 0.56 |
ENSDART00000138155
ENSDART00000079709 |
c6
|
complement component 6 |
| chr3_-_33422738 | 0.55 |
ENSDART00000075493
|
ccdc103
|
coiled-coil domain containing 103 |
| chr12_-_19282120 | 0.53 |
ENSDART00000153260
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr1_-_14694809 | 0.52 |
ENSDART00000193331
|
CU138548.5
|
|
| chr9_-_42696408 | 0.51 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr9_-_48397702 | 0.50 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr4_+_72235562 | 0.49 |
ENSDART00000168547
|
si:cabz01071912.2
|
si:cabz01071912.2 |
| chr22_+_17248145 | 0.49 |
ENSDART00000136908
|
axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr21_-_2958422 | 0.49 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr1_-_22851481 | 0.48 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr25_-_7753207 | 0.48 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr17_-_6382392 | 0.48 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr13_+_36633355 | 0.47 |
ENSDART00000135612
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr5_-_71705191 | 0.47 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr17_+_24613255 | 0.46 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr14_-_33351103 | 0.46 |
ENSDART00000010019
|
upf3b
|
UPF3B, regulator of nonsense mediated mRNA decay |
| chr20_-_22778394 | 0.46 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr3_+_31621774 | 0.46 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr24_+_12690163 | 0.45 |
ENSDART00000100644
|
rec8b
|
REC8 meiotic recombination protein b |
| chr4_+_22480169 | 0.45 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr3_-_16289826 | 0.45 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr13_+_27951688 | 0.45 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr12_+_18533198 | 0.45 |
ENSDART00000189729
|
meiob
|
meiosis specific with OB domains |
| chr9_-_30555725 | 0.44 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr17_+_33719415 | 0.44 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr5_+_42402536 | 0.43 |
ENSDART00000186754
|
BX548073.11
|
|
| chr17_+_14784275 | 0.43 |
ENSDART00000005355
|
rtraf
|
RNA transcription, translation and transport factor |
| chr5_-_34185115 | 0.42 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr19_-_25464291 | 0.42 |
ENSDART00000112915
|
umad1
|
UBAP1-MVB12-associated (UMA) domain containing 1 |
| chr25_+_30298377 | 0.42 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr24_+_34069675 | 0.42 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr4_-_8611841 | 0.41 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr23_+_20689255 | 0.41 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr5_-_66160415 | 0.41 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr6_+_49053319 | 0.41 |
ENSDART00000124524
|
sycp1
|
synaptonemal complex protein 1 |
| chr22_+_7439186 | 0.41 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr3_-_55139127 | 0.39 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr5_-_10007897 | 0.39 |
ENSDART00000109052
|
CR936408.1
|
Danio rerio uncharacterized LOC799523 (LOC799523), mRNA. |
| chr8_+_24747865 | 0.39 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr24_+_5840258 | 0.38 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr5_-_26181863 | 0.38 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr7_+_24049776 | 0.38 |
ENSDART00000166559
|
efs
|
embryonal Fyn-associated substrate |
| chr24_-_24038800 | 0.38 |
ENSDART00000080549
|
lyz
|
lysozyme |
| chr8_-_13972626 | 0.38 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr14_+_39255437 | 0.37 |
ENSDART00000147443
|
diaph2
|
diaphanous-related formin 2 |
| chr21_+_44557006 | 0.37 |
ENSDART00000170159
|
cmc4
|
C-x(9)-C motif containing 4 homolog (S. cerevisiae) |
| chr6_+_32834760 | 0.37 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr10_-_28761454 | 0.36 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr22_-_36934040 | 0.36 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr23_-_30045661 | 0.36 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr12_+_18532866 | 0.36 |
ENSDART00000152126
ENSDART00000152443 ENSDART00000089589 |
meiob
|
meiosis specific with OB domains |
| chr6_-_31364475 | 0.36 |
ENSDART00000145715
ENSDART00000134370 |
ak4
|
adenylate kinase 4 |
| chr12_+_35654749 | 0.35 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr8_+_42629748 | 0.35 |
ENSDART00000075553
ENSDART00000135525 |
crybb2
|
crystallin, beta B2 |
| chr6_-_6993046 | 0.34 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr6_-_40771813 | 0.34 |
ENSDART00000076200
|
h1fx
|
H1 histone family, member X |
| chr23_-_19486571 | 0.34 |
ENSDART00000009092
|
fam208ab
|
family with sequence similarity 208, member Ab |
| chr2_+_4383061 | 0.34 |
ENSDART00000163986
|
wacb
|
WW domain containing adaptor with coiled-coil b |
| chr21_+_13205859 | 0.33 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr16_-_5721386 | 0.33 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr20_-_44496245 | 0.33 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr12_-_684200 | 0.33 |
ENSDART00000152122
|
si:ch211-176g6.2
|
si:ch211-176g6.2 |
| chr11_-_7261717 | 0.33 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr2_-_24898226 | 0.33 |
ENSDART00000134936
|
si:dkey-149i17.9
|
si:dkey-149i17.9 |
| chr22_-_26945493 | 0.32 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr14_+_16765992 | 0.32 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
| chr17_+_35431724 | 0.32 |
ENSDART00000190293
|
BX571665.1
|
|
| chr16_+_33902006 | 0.32 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr23_+_23658474 | 0.32 |
ENSDART00000162838
|
agrn
|
agrin |
| chr8_+_46536893 | 0.32 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr21_-_40782393 | 0.32 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr17_+_31592191 | 0.31 |
ENSDART00000153765
|
si:dkey-13p1.3
|
si:dkey-13p1.3 |
| chr7_+_39706004 | 0.31 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr4_-_17642168 | 0.31 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr18_+_30508729 | 0.31 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr18_-_26101800 | 0.31 |
ENSDART00000004692
|
idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr13_+_15682803 | 0.31 |
ENSDART00000188063
|
CR931980.1
|
|
| chr21_+_19858627 | 0.31 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr10_-_27566481 | 0.31 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr8_+_23731483 | 0.31 |
ENSDART00000099751
|
fance
|
Fanconi anemia, complementation group E |
| chr25_+_19739665 | 0.30 |
ENSDART00000067353
|
zgc:101783
|
zgc:101783 |
| chr24_-_26399623 | 0.30 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr25_+_4954220 | 0.30 |
ENSDART00000156034
|
si:ch73-265h17.2
|
si:ch73-265h17.2 |
| chr22_+_18530395 | 0.30 |
ENSDART00000105415
ENSDART00000183958 |
si:ch211-212d10.1
|
si:ch211-212d10.1 |
| chr10_+_39084354 | 0.30 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr11_-_44876005 | 0.29 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr16_+_32092866 | 0.29 |
ENSDART00000137785
|
si:dkey-40m6.11
|
si:dkey-40m6.11 |
| chr16_-_23800484 | 0.29 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr7_-_59256806 | 0.29 |
ENSDART00000167955
|
m1ap
|
meiosis 1 associated protein |
| chr14_-_33218418 | 0.29 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr21_-_20929575 | 0.29 |
ENSDART00000163889
|
c6
|
complement component 6 |
| chr22_+_35068046 | 0.29 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr11_-_8262105 | 0.29 |
ENSDART00000173225
|
si:cabz01021066.1
|
si:cabz01021066.1 |
| chr9_+_42066030 | 0.28 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr5_+_28160503 | 0.28 |
ENSDART00000051516
|
tacr1a
|
tachykinin receptor 1a |
| chr2_+_49580450 | 0.28 |
ENSDART00000056252
|
sema4e
|
semaphorin 4e |
| chr22_-_31752937 | 0.28 |
ENSDART00000169611
|
CABZ01028298.1
|
|
| chr17_+_16873417 | 0.28 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr21_+_15713097 | 0.28 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr7_+_19903924 | 0.28 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr5_+_28161079 | 0.28 |
ENSDART00000141109
|
tacr1a
|
tachykinin receptor 1a |
| chr19_-_10243148 | 0.28 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr5_+_28313824 | 0.27 |
ENSDART00000135878
|
si:dkeyp-86b9.1
|
si:dkeyp-86b9.1 |
| chr4_-_11577253 | 0.27 |
ENSDART00000144452
|
net1
|
neuroepithelial cell transforming 1 |
| chr12_+_26471712 | 0.27 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr18_-_21929426 | 0.27 |
ENSDART00000132034
ENSDART00000079050 |
nutf2
|
nuclear transport factor 2 |
| chr2_-_41861040 | 0.27 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr19_+_41464870 | 0.27 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr5_+_37087583 | 0.27 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr2_-_29761965 | 0.27 |
ENSDART00000142057
ENSDART00000110595 |
si:dkey-188g12.1
|
si:dkey-188g12.1 |
| chr11_-_29657947 | 0.27 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr14_+_5861435 | 0.27 |
ENSDART00000041279
ENSDART00000147341 |
tubb4b
|
tubulin, beta 4B class IVb |
| chr12_+_13256415 | 0.27 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr5_-_9073433 | 0.26 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr21_-_20832482 | 0.26 |
ENSDART00000191928
|
c6
|
complement component 6 |
| chr16_-_38553721 | 0.26 |
ENSDART00000109124
|
rspo2
|
R-spondin 2 |
| chr12_-_6880694 | 0.26 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr9_+_38399912 | 0.26 |
ENSDART00000022246
ENSDART00000145892 |
bcs1l
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr23_+_36653376 | 0.26 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr23_-_16682186 | 0.26 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr21_-_5879897 | 0.26 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr20_+_3934516 | 0.26 |
ENSDART00000165732
|
clec11a
|
C-type lectin domain containing 11A |
| chr4_-_16412084 | 0.26 |
ENSDART00000188460
|
dcn
|
decorin |
| chr22_+_19552987 | 0.25 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr17_+_43889371 | 0.25 |
ENSDART00000156871
ENSDART00000154702 |
msh4
|
mutS homolog 4 |
| chr4_-_18635005 | 0.25 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr22_+_19553390 | 0.25 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr7_+_22688781 | 0.24 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr11_-_29658396 | 0.24 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr13_+_13821054 | 0.24 |
ENSDART00000127417
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr7_-_51321126 | 0.24 |
ENSDART00000067647
|
rasl11a
|
RAS-like, family 11, member A |
| chr2_+_31600661 | 0.24 |
ENSDART00000139039
|
si:ch211-106h4.4
|
si:ch211-106h4.4 |
| chr11_-_3865472 | 0.24 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr17_+_42842632 | 0.24 |
ENSDART00000155547
ENSDART00000126087 |
qpct
|
glutaminyl-peptide cyclotransferase |
| chr10_-_17179415 | 0.24 |
ENSDART00000131751
|
depdc5
|
DEP domain containing 5 |
| chr15_-_21837207 | 0.24 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr21_+_11503212 | 0.24 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr11_-_31039533 | 0.23 |
ENSDART00000127355
|
ier2b
|
immediate early response 2b |
| chr15_-_23645810 | 0.23 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr2_+_10134345 | 0.23 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr6_+_13920479 | 0.23 |
ENSDART00000155480
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr14_+_28438947 | 0.23 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr9_-_2572790 | 0.23 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr20_-_12642685 | 0.23 |
ENSDART00000173257
|
si:dkey-97l20.6
|
si:dkey-97l20.6 |
| chr7_+_19904136 | 0.23 |
ENSDART00000173452
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr6_+_25261297 | 0.23 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr1_+_22851261 | 0.23 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr3_-_28750495 | 0.23 |
ENSDART00000054408
|
gsg1l
|
gsg1-like |
| chr23_+_21663631 | 0.23 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr5_-_48268049 | 0.23 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr1_+_17376922 | 0.23 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr13_+_35635672 | 0.23 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr22_-_24285432 | 0.23 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr12_+_26632448 | 0.23 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr13_+_33024547 | 0.23 |
ENSDART00000057377
|
arg2
|
arginase 2 |
| chr1_+_2129164 | 0.22 |
ENSDART00000074923
ENSDART00000124534 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr6_-_7082108 | 0.22 |
ENSDART00000081760
|
ihhb
|
Indian hedgehog homolog b |
| chr13_-_17723417 | 0.22 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr8_-_20138054 | 0.22 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr11_-_6265574 | 0.22 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr20_+_18232893 | 0.22 |
ENSDART00000189225
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr2_+_16460321 | 0.22 |
ENSDART00000145107
|
agfg1b
|
ArfGAP with FG repeats 1b |
| chr2_-_16159491 | 0.22 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr5_+_37837245 | 0.22 |
ENSDART00000171617
|
epd
|
ependymin |
| chr17_-_19345521 | 0.22 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr5_-_30984271 | 0.22 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr8_-_15292197 | 0.22 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr5_-_10239079 | 0.22 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr17_+_46739693 | 0.22 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr11_+_13630107 | 0.22 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr10_-_10864331 | 0.22 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr15_-_21837515 | 0.21 |
ENSDART00000185423
|
sik2b
|
salt-inducible kinase 2b |
| chr17_+_33453689 | 0.21 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr14_+_3522334 | 0.21 |
ENSDART00000164547
|
fnta
|
farnesyltransferase, CAAX box, alpha |
| chr21_-_43474012 | 0.21 |
ENSDART00000065104
|
tmem185
|
transmembrane protein 185 |
| chr15_-_28223757 | 0.21 |
ENSDART00000110969
ENSDART00000138401 |
scarf1
|
scavenger receptor class F, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfia
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.2 | 2.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.6 | GO:0071314 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.2 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.7 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.2 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.5 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.4 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.8 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.3 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 1.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0033335 | anal fin development(GO:0033335) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.2 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.2 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.3 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.7 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 2.3 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.5 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.2 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0060307 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell action potential(GO:0086005) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.0 | 0.1 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.1 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0051230 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 1.0 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.4 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0048664 | auditory receptor cell fate commitment(GO:0009912) neuron fate determination(GO:0048664) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.9 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 0.0 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.0 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0060846 | blood vessel endothelial cell fate commitment(GO:0060846) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 2.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 1.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 1.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.2 | 0.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 0.6 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.2 | 0.7 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.9 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 3.6 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.2 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0046977 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.0 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 0.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |