Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
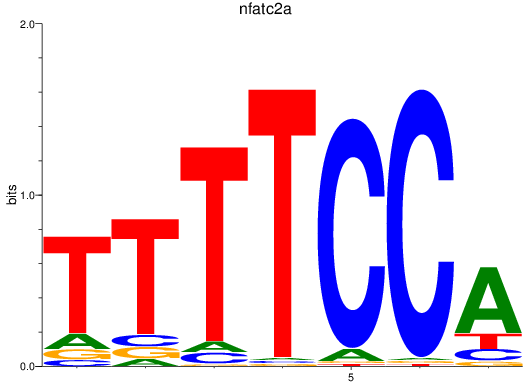
Results for nfatc2a
Z-value: 1.24
Transcription factors associated with nfatc2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc2a
|
ENSDARG00000100927 | nuclear factor of activated T cells 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc2a | dr11_v1_chr23_+_39089574_39089574 | -0.55 | 1.7e-02 | Click! |
Activity profile of nfatc2a motif
Sorted Z-values of nfatc2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_46040666 | 1.41 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr19_+_41479990 | 1.27 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr24_-_17029374 | 1.20 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr12_-_25887864 | 1.15 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr5_-_3991655 | 1.11 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr17_-_30521043 | 1.11 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr8_+_13106760 | 1.08 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr10_+_5954787 | 1.06 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr7_+_33424044 | 1.04 |
ENSDART00000180260
|
glceb
|
glucuronic acid epimerase b |
| chr18_-_39696853 | 1.01 |
ENSDART00000148383
|
dmxl2
|
Dmx-like 2 |
| chr1_+_44173506 | 0.99 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr23_-_33558161 | 0.98 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr1_+_14454663 | 0.97 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr4_-_1720648 | 0.97 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr8_-_22558773 | 0.90 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr2_+_24638367 | 0.89 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr6_+_36821621 | 0.89 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr1_+_36771954 | 0.88 |
ENSDART00000149022
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr3_-_15470944 | 0.88 |
ENSDART00000185302
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr5_-_69041102 | 0.87 |
ENSDART00000161561
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr11_+_24046179 | 0.87 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr14_+_26759332 | 0.87 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr16_+_12812472 | 0.87 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr11_-_21404358 | 0.86 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr6_-_39313027 | 0.86 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr23_+_27782071 | 0.85 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr22_-_26353916 | 0.84 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr14_+_6615564 | 0.83 |
ENSDART00000139292
|
si:dkeyp-44a8.2
|
si:dkeyp-44a8.2 |
| chr19_-_4137087 | 0.80 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr16_-_13281380 | 0.78 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr6_-_49547680 | 0.78 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr1_+_52632856 | 0.77 |
ENSDART00000011725
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr17_+_20569806 | 0.76 |
ENSDART00000113936
|
SAMD8
|
zgc:162183 |
| chr1_+_26677406 | 0.75 |
ENSDART00000183427
ENSDART00000180366 ENSDART00000181997 |
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr9_-_23807032 | 0.75 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr11_-_11882982 | 0.75 |
ENSDART00000190853
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr11_-_33618612 | 0.73 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_+_6938289 | 0.73 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr9_-_11263228 | 0.73 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr23_+_1181248 | 0.73 |
ENSDART00000170942
|
utrn
|
utrophin |
| chr12_-_3133483 | 0.72 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr13_-_24825691 | 0.71 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr9_-_9209831 | 0.71 |
ENSDART00000026275
|
cbsb
|
cystathionine-beta-synthase b |
| chr21_+_15756549 | 0.70 |
ENSDART00000026903
|
ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr16_-_6205790 | 0.70 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr4_-_3353595 | 0.70 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_+_26676758 | 0.69 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr8_+_47683539 | 0.69 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr16_-_38333976 | 0.69 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr14_+_31509922 | 0.69 |
ENSDART00000124499
|
hprt1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr19_-_6193448 | 0.69 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr2_+_15069011 | 0.68 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr10_+_44986419 | 0.68 |
ENSDART00000162566
|
h2afvb
|
H2A histone family, member Vb |
| chr13_-_40282770 | 0.68 |
ENSDART00000140875
|
zgc:123010
|
zgc:123010 |
| chr19_-_24267410 | 0.68 |
ENSDART00000104083
|
s100v2
|
S100 calcium binding protein V2 |
| chr8_+_47683352 | 0.68 |
ENSDART00000187320
ENSDART00000192605 |
dpp9
|
dipeptidyl-peptidase 9 |
| chr1_-_43920371 | 0.68 |
ENSDART00000109283
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr5_-_19963537 | 0.68 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr10_-_41352502 | 0.67 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr2_+_35993404 | 0.67 |
ENSDART00000170845
|
lamc2
|
laminin, gamma 2 |
| chr18_+_50669456 | 0.67 |
ENSDART00000163005
|
si:dkey-151j17.4
|
si:dkey-151j17.4 |
| chr12_-_3840664 | 0.67 |
ENSDART00000160967
|
taok2b
|
TAO kinase 2b |
| chr5_+_31168096 | 0.66 |
ENSDART00000086443
ENSDART00000192271 |
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr20_-_18789543 | 0.66 |
ENSDART00000182240
|
ccm2
|
cerebral cavernous malformation 2 |
| chr16_-_17345377 | 0.66 |
ENSDART00000143056
|
zyx
|
zyxin |
| chr3_-_20063671 | 0.66 |
ENSDART00000056614
ENSDART00000126915 |
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr17_-_11418513 | 0.66 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr20_+_14977260 | 0.66 |
ENSDART00000186424
|
vamp4
|
vesicle-associated membrane protein 4 |
| chr17_-_8592824 | 0.65 |
ENSDART00000127022
|
CU462878.1
|
|
| chr7_+_51795667 | 0.65 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr9_-_16133263 | 0.65 |
ENSDART00000077187
|
myo1b
|
myosin IB |
| chr5_-_68074592 | 0.65 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr24_-_34335265 | 0.64 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr21_+_6291027 | 0.64 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr4_-_7876005 | 0.64 |
ENSDART00000109252
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr15_-_8856785 | 0.64 |
ENSDART00000192816
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr23_+_9522942 | 0.64 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr19_+_46222918 | 0.63 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr24_-_32025637 | 0.63 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr22_+_15720381 | 0.63 |
ENSDART00000128149
|
fam32a
|
family with sequence similarity 32, member A |
| chr3_-_37699992 | 0.62 |
ENSDART00000151193
|
gpatch8
|
G patch domain containing 8 |
| chr10_+_44955106 | 0.62 |
ENSDART00000185837
|
il1b
|
interleukin 1, beta |
| chr16_-_21903083 | 0.62 |
ENSDART00000165849
|
setdb1b
|
SET domain, bifurcated 1b |
| chr12_+_17603528 | 0.62 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr9_+_24936496 | 0.62 |
ENSDART00000157474
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr3_-_40054615 | 0.62 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr19_-_20403507 | 0.62 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr5_-_48664522 | 0.62 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr14_-_46616487 | 0.62 |
ENSDART00000105417
ENSDART00000166550 ENSDART00000105418 |
prom1a
|
prominin 1a |
| chr12_+_33403694 | 0.62 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr13_+_13681681 | 0.61 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr17_-_45370200 | 0.61 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr25_-_25566363 | 0.61 |
ENSDART00000185770
|
hic1l
|
hypermethylated in cancer 1 like |
| chr19_-_11336782 | 0.61 |
ENSDART00000131014
|
sept7a
|
septin 7a |
| chr9_-_12659140 | 0.61 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr19_+_43684376 | 0.60 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr22_+_17433968 | 0.60 |
ENSDART00000139523
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr9_+_16241656 | 0.60 |
ENSDART00000154326
|
si:ch211-261p9.4
|
si:ch211-261p9.4 |
| chr16_-_42461263 | 0.59 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr17_+_21887823 | 0.59 |
ENSDART00000131929
ENSDART00000165192 |
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr16_-_7793457 | 0.59 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr2_-_37353098 | 0.59 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr14_-_31856819 | 0.59 |
ENSDART00000003345
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr7_+_13609457 | 0.59 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr9_-_19092322 | 0.59 |
ENSDART00000131957
|
crfb2
|
cytokine receptor family member b2 |
| chr3_-_40976288 | 0.59 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr3_-_23575007 | 0.58 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr15_+_21262917 | 0.58 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr6_+_10333920 | 0.58 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr19_-_46058963 | 0.58 |
ENSDART00000170409
|
nup153
|
nucleoporin 153 |
| chr17_-_15528597 | 0.58 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr21_-_2814709 | 0.57 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr20_-_2355357 | 0.57 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr3_+_40409100 | 0.57 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr11_+_10909183 | 0.57 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr25_-_3745393 | 0.57 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr12_+_22560067 | 0.57 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr5_-_40190949 | 0.57 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr5_+_13521081 | 0.56 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr17_+_25519089 | 0.56 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr3_-_5067585 | 0.56 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr19_-_20403845 | 0.56 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr10_-_6867282 | 0.56 |
ENSDART00000144001
ENSDART00000109744 |
ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr19_+_37120491 | 0.56 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr12_-_3453589 | 0.56 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr21_+_37090585 | 0.56 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr9_+_33158191 | 0.56 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr16_+_23397785 | 0.56 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr23_+_27779452 | 0.55 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr6_+_27667359 | 0.55 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr15_-_29354020 | 0.55 |
ENSDART00000127795
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr21_+_37090795 | 0.55 |
ENSDART00000085786
|
znf346
|
zinc finger protein 346 |
| chr8_+_53423408 | 0.54 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr2_+_36015049 | 0.54 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr25_-_36263115 | 0.54 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr23_+_19790962 | 0.54 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr14_+_45028062 | 0.54 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr16_+_12812214 | 0.54 |
ENSDART00000124875
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr22_-_21046843 | 0.53 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr16_-_48428035 | 0.53 |
ENSDART00000190554
|
rad21a
|
RAD21 cohesin complex component a |
| chr19_+_9050852 | 0.53 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr20_-_2298970 | 0.53 |
ENSDART00000136067
|
EPB41L2
|
si:ch73-18b11.1 |
| chr23_+_1276006 | 0.53 |
ENSDART00000162294
|
utrn
|
utrophin |
| chr3_-_46410387 | 0.53 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr2_-_3077027 | 0.53 |
ENSDART00000109319
|
arf1
|
ADP-ribosylation factor 1 |
| chr6_+_37754763 | 0.53 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr25_-_3623847 | 0.53 |
ENSDART00000172586
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr6_-_19333947 | 0.53 |
ENSDART00000160887
|
gga3
|
ggolgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr23_-_24682244 | 0.52 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr21_-_3700334 | 0.52 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr19_-_6193067 | 0.52 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr3_+_53156813 | 0.52 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr23_-_18415872 | 0.52 |
ENSDART00000135430
|
fam120c
|
family with sequence similarity 120C |
| chr15_+_19991280 | 0.52 |
ENSDART00000186677
|
zgc:112083
|
zgc:112083 |
| chr6_-_49537646 | 0.52 |
ENSDART00000180438
|
FO704848.1
|
|
| chr18_+_27489595 | 0.52 |
ENSDART00000182018
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr10_+_36029537 | 0.51 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr21_+_18405585 | 0.51 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr13_+_33024547 | 0.51 |
ENSDART00000057377
|
arg2
|
arginase 2 |
| chr8_+_28452738 | 0.51 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr13_+_25364753 | 0.51 |
ENSDART00000027428
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr19_-_27827744 | 0.51 |
ENSDART00000181620
|
papd7
|
PAP associated domain containing 7 |
| chr13_-_42749916 | 0.50 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr14_-_22495604 | 0.50 |
ENSDART00000137167
|
si:ch211-107m4.1
|
si:ch211-107m4.1 |
| chr8_-_38022298 | 0.50 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr11_+_5588122 | 0.50 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr20_-_45807982 | 0.50 |
ENSDART00000074546
|
fermt1
|
fermitin family member 1 |
| chr10_-_1718395 | 0.50 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr12_-_23320266 | 0.50 |
ENSDART00000181711
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr3_+_30921246 | 0.50 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr22_-_20950448 | 0.50 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr8_-_33154677 | 0.50 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr1_+_44173245 | 0.49 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr17_+_25444323 | 0.49 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr12_+_16953415 | 0.49 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr20_+_6535176 | 0.49 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr5_+_22133153 | 0.49 |
ENSDART00000016214
|
msna
|
moesin a |
| chr8_-_9570511 | 0.49 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr25_+_26901149 | 0.49 |
ENSDART00000153839
|
znf800a
|
zinc finger protein 800a |
| chr8_+_49975160 | 0.49 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr6_-_7735153 | 0.49 |
ENSDART00000151545
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr21_-_43482426 | 0.49 |
ENSDART00000192901
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr20_-_3319642 | 0.49 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr9_-_9842149 | 0.48 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr2_-_38206034 | 0.48 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr21_+_31253048 | 0.48 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr23_-_18668836 | 0.48 |
ENSDART00000138792
ENSDART00000051182 |
arhgap4b
|
Rho GTPase activating protein 4b |
| chr25_-_12902242 | 0.47 |
ENSDART00000164733
|
sept15
|
septin 15 |
| chr13_+_25505580 | 0.47 |
ENSDART00000140634
|
inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr19_-_13774502 | 0.47 |
ENSDART00000159711
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr20_+_27087539 | 0.47 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr22_+_17536989 | 0.47 |
ENSDART00000149531
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr2_-_37462462 | 0.47 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr10_-_3258073 | 0.47 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr13_+_39297802 | 0.47 |
ENSDART00000133636
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr9_+_37366973 | 0.47 |
ENSDART00000016370
|
dirc2
|
disrupted in renal carcinoma 2 |
| chr22_-_21046654 | 0.47 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr20_-_14462995 | 0.47 |
ENSDART00000152418
ENSDART00000044125 |
grcc10
|
gene rich cluster, C10 gene |
| chr9_+_23003208 | 0.47 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr19_-_10214264 | 0.47 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfatc2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.3 | 1.2 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 0.9 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.3 | 1.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 0.8 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.3 | 1.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.7 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.2 | 0.7 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 0.7 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.2 | 0.7 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.2 | 0.6 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.2 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.2 | 0.5 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 1.0 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.2 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.0 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.5 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.2 | 0.6 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 1.4 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.6 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.9 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.6 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.4 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.5 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.5 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.6 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.5 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.1 | 0.9 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 0.4 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.9 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.1 | 0.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 1.0 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.2 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.3 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.3 | GO:0071962 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.5 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.4 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.2 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.2 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.4 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.2 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.1 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.4 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.1 | 0.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.2 | GO:0032241 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.2 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.1 | 0.6 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.2 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.8 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.6 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 0.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.2 | GO:0034154 | toll-like receptor 3 signaling pathway(GO:0034138) toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.7 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.1 | 0.3 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 0.4 | GO:0030643 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.0 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.2 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.3 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.6 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.2 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.3 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.0 | 1.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0033337 | dorsal fin development(GO:0033337) |
| 0.0 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 1.5 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.6 | GO:0006984 | ER-nucleus signaling pathway(GO:0006984) |
| 0.0 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0046639 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) negative regulation of T cell differentiation(GO:0045581) regulation of T-helper cell differentiation(GO:0045622) negative regulation of T-helper cell differentiation(GO:0045623) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) negative regulation of alpha-beta T cell differentiation(GO:0046639) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.5 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.3 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.3 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 1.3 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.4 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0051785 | positive regulation of mitotic nuclear division(GO:0045840) positive regulation of nuclear division(GO:0051785) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.9 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.7 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament organization(GO:0045109) intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.0 | GO:0048883 | neuromast primordium migration(GO:0048883) |
| 0.0 | 0.5 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.6 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 1.0 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.7 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 1.1 | GO:0051236 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.6 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.4 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:0007309 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte animal/vegetal axis specification(GO:0060832) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 1.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 1.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 1.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.1 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.2 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 3.3 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.3 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.2 | GO:0005923 | bicellular tight junction(GO:0005923) apical junction complex(GO:0043296) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.3 | 1.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 0.9 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.3 | 1.0 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.2 | 0.7 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.2 | 1.8 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 0.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.6 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.2 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.9 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.2 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.6 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.0 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.7 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.6 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.5 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.9 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.3 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.7 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 2.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 2.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.3 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 9.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 2.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 2.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 3.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.6 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME AXON GUIDANCE | Genes involved in Axon guidance |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |