Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
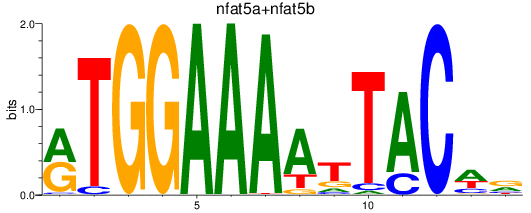
Results for nfat5a+nfat5b
Z-value: 0.20
Transcription factors associated with nfat5a+nfat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfat5a
|
ENSDARG00000020872 | nuclear factor of activated T cells 5a |
|
nfat5b
|
ENSDARG00000102556 | nuclear factor of activated T cells 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfat5a | dr11_v1_chr25_-_37121335_37121335 | 0.57 | 1.4e-02 | Click! |
| nfat5b | dr11_v1_chr7_+_67381912_67381912 | 0.37 | 1.4e-01 | Click! |
Activity profile of nfat5a+nfat5b motif
Sorted Z-values of nfat5a+nfat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_70271799 | 1.14 |
ENSDART00000101316
|
znf618
|
zinc finger protein 618 |
| chr2_-_38284648 | 0.64 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr10_+_15255012 | 0.49 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr6_+_612330 | 0.49 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr15_-_25099679 | 0.47 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr16_+_2469889 | 0.43 |
ENSDART00000126759
|
cnih4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr23_+_2728095 | 0.43 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr23_-_31645760 | 0.41 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_25099233 | 0.37 |
ENSDART00000192617
|
rflnb
|
refilin B |
| chr6_-_19381993 | 0.37 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr16_+_43077909 | 0.32 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr19_-_25149034 | 0.31 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr7_+_55950229 | 0.31 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr5_+_13521081 | 0.31 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr9_-_46415847 | 0.30 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr15_+_11693624 | 0.30 |
ENSDART00000193630
ENSDART00000161930 |
strn4
|
striatin, calmodulin binding protein 4 |
| chr24_-_19719240 | 0.29 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr14_-_34633960 | 0.28 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr6_-_12900154 | 0.28 |
ENSDART00000080408
ENSDART00000150887 |
ical1
|
islet cell autoantigen 1-like |
| chr9_+_23003208 | 0.27 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr15_+_21707900 | 0.27 |
ENSDART00000141948
|
NKAPD1
|
zgc:162339 |
| chr18_-_13133268 | 0.27 |
ENSDART00000081811
|
zgc:112052
|
zgc:112052 |
| chr2_+_36616830 | 0.24 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr19_-_3056235 | 0.23 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr1_+_19616799 | 0.23 |
ENSDART00000143077
|
fbxo10
|
F-box protein 10 |
| chr22_-_8306743 | 0.23 |
ENSDART00000123982
|
CABZ01077218.1
|
|
| chr20_-_487783 | 0.22 |
ENSDART00000162218
|
col10a1b
|
collagen, type X, alpha 1b |
| chr19_-_4785734 | 0.22 |
ENSDART00000113088
|
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr15_-_18209672 | 0.22 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr21_+_21279159 | 0.22 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr19_+_15440841 | 0.21 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_-_6115688 | 0.21 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr24_-_24060460 | 0.20 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr12_-_17147473 | 0.19 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr1_-_8566567 | 0.18 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr5_+_42259002 | 0.18 |
ENSDART00000083778
|
eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr13_+_25364324 | 0.18 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr1_-_53685090 | 0.18 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr17_+_15559046 | 0.18 |
ENSDART00000187126
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr21_-_2814709 | 0.17 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr2_+_29996650 | 0.17 |
ENSDART00000138050
ENSDART00000141026 |
rbm33b
|
RNA binding motif protein 33b |
| chr24_-_24060632 | 0.17 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr17_+_33415319 | 0.17 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr7_+_29115890 | 0.16 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr22_+_4488454 | 0.16 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr19_+_32807236 | 0.16 |
ENSDART00000008698
|
stk3
|
serine/threonine kinase 3 (STE20 homolog, yeast) |
| chr9_+_7909490 | 0.16 |
ENSDART00000133063
ENSDART00000109288 |
myo16
|
myosin XVI |
| chr23_-_36439961 | 0.16 |
ENSDART00000187907
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr17_-_33412868 | 0.16 |
ENSDART00000187521
|
BX323819.1
|
|
| chr13_+_1015749 | 0.15 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr6_+_6802142 | 0.15 |
ENSDART00000065566
|
dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr18_+_16715864 | 0.15 |
ENSDART00000079758
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr5_-_40040488 | 0.15 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr10_-_32465462 | 0.15 |
ENSDART00000134056
|
uvrag
|
UV radiation resistance associated gene |
| chr21_+_31253048 | 0.15 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr1_+_54124209 | 0.15 |
ENSDART00000187730
|
LO017722.1
|
|
| chr3_+_50172452 | 0.15 |
ENSDART00000191224
|
epn3a
|
epsin 3a |
| chr17_-_33414781 | 0.15 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr1_+_2712956 | 0.14 |
ENSDART00000126093
|
gpc6a
|
glypican 6a |
| chr19_+_43885770 | 0.14 |
ENSDART00000135599
|
lypla2
|
lysophospholipase II |
| chr9_+_33145522 | 0.14 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr17_+_33415542 | 0.14 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr3_-_5067585 | 0.14 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr9_+_34319333 | 0.14 |
ENSDART00000035522
ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr20_+_400487 | 0.13 |
ENSDART00000108872
|
ndufaf4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr1_-_493218 | 0.13 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr2_-_42492201 | 0.13 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr11_+_11751006 | 0.13 |
ENSDART00000180372
ENSDART00000081018 |
jupa
|
junction plakoglobin a |
| chr10_+_15511885 | 0.12 |
ENSDART00000165663
|
erbin
|
erbb2 interacting protein |
| chr17_-_36529449 | 0.12 |
ENSDART00000187252
|
colec11
|
collectin sub-family member 11 |
| chr3_+_32080403 | 0.12 |
ENSDART00000124943
|
aldh16a1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr9_+_6255682 | 0.11 |
ENSDART00000149827
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr5_-_62940851 | 0.11 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr17_-_36529016 | 0.11 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr16_+_45746549 | 0.10 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr17_+_43013171 | 0.09 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr2_-_42492445 | 0.09 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr6_+_40992883 | 0.09 |
ENSDART00000076061
|
tgfa
|
transforming growth factor, alpha |
| chr1_-_23595779 | 0.09 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr18_+_41772474 | 0.09 |
ENSDART00000097547
|
trpc6b
|
transient receptor potential cation channel, subfamily C, member 6b |
| chr8_+_1843135 | 0.09 |
ENSDART00000141452
|
snap29
|
synaptosomal-associated protein 29 |
| chr25_+_30131055 | 0.09 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr18_+_26895994 | 0.08 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr17_-_277046 | 0.08 |
ENSDART00000182587
|
LO018437.1
|
|
| chr13_-_42749916 | 0.08 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr8_-_4789034 | 0.08 |
ENSDART00000082256
|
ciao1
|
cytosolic iron-sulfur assembly component 1 |
| chr1_+_54194801 | 0.08 |
ENSDART00000186802
|
CABZ01067150.1
|
|
| chr5_+_1515938 | 0.08 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr22_+_1947494 | 0.07 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr9_+_6255973 | 0.07 |
ENSDART00000078525
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr13_+_18321140 | 0.07 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr19_-_42416696 | 0.07 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr7_+_32021669 | 0.07 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr8_-_43677762 | 0.07 |
ENSDART00000167762
|
ep400
|
E1A binding protein p400 |
| chr2_-_26499842 | 0.07 |
ENSDART00000186929
|
BX569796.1
|
|
| chr13_-_41155472 | 0.07 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr24_-_20599781 | 0.07 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr12_+_34891529 | 0.06 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr10_+_44955106 | 0.06 |
ENSDART00000185837
|
il1b
|
interleukin 1, beta |
| chr1_+_44554105 | 0.06 |
ENSDART00000149764
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr12_-_30549022 | 0.05 |
ENSDART00000102474
|
zgc:158404
|
zgc:158404 |
| chr4_+_10604110 | 0.05 |
ENSDART00000122636
ENSDART00000037140 |
ing3
|
inhibitor of growth family, member 3 |
| chr8_-_45729248 | 0.05 |
ENSDART00000187736
|
BX088696.1
|
|
| chr1_+_56972616 | 0.05 |
ENSDART00000152812
|
si:ch211-1f22.11
|
si:ch211-1f22.11 |
| chr1_+_41666611 | 0.05 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr6_+_51932563 | 0.04 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
| chr5_-_13353810 | 0.04 |
ENSDART00000146136
ENSDART00000099602 ENSDART00000004539 |
ube2l3b
|
ubiquitin-conjugating enzyme E2L 3b |
| chr3_-_2033105 | 0.04 |
ENSDART00000135249
|
si:dkey-88j15.3
|
si:dkey-88j15.3 |
| chr25_+_18583877 | 0.04 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr5_-_9948497 | 0.04 |
ENSDART00000183196
|
tor4ab
|
torsin family 4, member Ab |
| chr10_+_5954787 | 0.04 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr9_+_310331 | 0.04 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr23_+_26102028 | 0.04 |
ENSDART00000139488
|
si:dkey-78k11.4
|
si:dkey-78k11.4 |
| chr16_-_21047872 | 0.03 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr7_+_39624728 | 0.03 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr7_-_32021853 | 0.03 |
ENSDART00000134521
|
kif18a
|
kinesin family member 18A |
| chr18_-_33085006 | 0.03 |
ENSDART00000182992
|
CABZ01032906.1
|
|
| chr19_-_10214264 | 0.03 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr22_-_9183042 | 0.03 |
ENSDART00000178327
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr25_+_7321675 | 0.03 |
ENSDART00000104712
ENSDART00000142934 |
hmg20a
|
high mobility group 20A |
| chr7_-_29115772 | 0.03 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr21_-_15282452 | 0.03 |
ENSDART00000143113
|
sfswap
|
splicing factor SWAP |
| chr23_-_45897900 | 0.03 |
ENSDART00000149263
|
ftr92
|
finTRIM family, member 92 |
| chr13_-_24874950 | 0.03 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr3_-_29688370 | 0.03 |
ENSDART00000151147
ENSDART00000151679 |
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr9_+_53725294 | 0.03 |
ENSDART00000165991
|
cnmd
|
chondromodulin |
| chr7_+_20645121 | 0.03 |
ENSDART00000052922
|
sat2a
|
spermidine/spermine N1-acetyltransferase family member 2a |
| chr20_+_10498704 | 0.03 |
ENSDART00000192985
|
zgc:100997
|
zgc:100997 |
| chr4_+_76722754 | 0.03 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr1_+_57041549 | 0.03 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr1_+_19303241 | 0.03 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr12_-_17602958 | 0.02 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr17_+_24756379 | 0.02 |
ENSDART00000185059
|
stac
|
SH3 and cysteine rich domain |
| chr20_+_46206998 | 0.02 |
ENSDART00000074482
|
taar15
|
trace amine associated receptor 15 |
| chr20_+_10498986 | 0.02 |
ENSDART00000064114
|
zgc:100997
|
zgc:100997 |
| chr2_-_5502256 | 0.02 |
ENSDART00000193037
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr21_+_5494561 | 0.02 |
ENSDART00000169728
|
ly6m3
|
lymphocyte antigen 6 family member M3 |
| chr4_-_53047883 | 0.02 |
ENSDART00000157496
|
si:dkey-201g16.5
|
si:dkey-201g16.5 |
| chr7_+_56098590 | 0.02 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr9_+_53725128 | 0.02 |
ENSDART00000169062
|
cnmd
|
chondromodulin |
| chr9_-_11263228 | 0.02 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr1_+_56502706 | 0.02 |
ENSDART00000188665
|
CABZ01059403.1
|
|
| chr13_+_22104298 | 0.02 |
ENSDART00000115393
|
si:dkey-174i8.1
|
si:dkey-174i8.1 |
| chr4_-_58883559 | 0.02 |
ENSDART00000164546
|
si:dkey-28i19.4
|
si:dkey-28i19.4 |
| chr19_+_29854223 | 0.02 |
ENSDART00000109729
ENSDART00000157419 |
si:ch73-130a3.4
|
si:ch73-130a3.4 |
| chr23_-_27875140 | 0.02 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr4_+_47751483 | 0.02 |
ENSDART00000160043
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr18_-_898870 | 0.02 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr10_+_13209580 | 0.02 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr2_+_3881000 | 0.02 |
ENSDART00000081897
|
mpp7b
|
membrane protein, palmitoylated 7b (MAGUK p55 subfamily member 7) |
| chr18_+_9641210 | 0.02 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr3_-_1906976 | 0.02 |
ENSDART00000180953
|
zgc:101806
|
zgc:101806 |
| chr16_+_12660477 | 0.02 |
ENSDART00000016834
|
aicda
|
activation-induced cytidine deaminase |
| chr5_+_53479576 | 0.02 |
ENSDART00000099616
ENSDART00000169169 |
BX323994.1
|
|
| chr6_-_42073367 | 0.01 |
ENSDART00000154304
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr2_-_29996036 | 0.01 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr6_-_57635577 | 0.01 |
ENSDART00000168708
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr2_-_34483597 | 0.01 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr9_+_1352603 | 0.01 |
ENSDART00000146016
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr7_+_57795974 | 0.01 |
ENSDART00000148369
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr20_-_53624645 | 0.01 |
ENSDART00000083427
ENSDART00000152920 |
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr10_-_37072776 | 0.01 |
ENSDART00000110835
|
myo18aa
|
myosin XVIIIAa |
| chr7_+_29009161 | 0.01 |
ENSDART00000166458
|
CR376738.1
|
|
| chr14_-_30945515 | 0.01 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr9_-_43082945 | 0.01 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr21_+_6291027 | 0.01 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr5_-_37900350 | 0.01 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr19_-_36234185 | 0.01 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr9_+_30478768 | 0.00 |
ENSDART00000101097
|
acp6
|
acid phosphatase 6, lysophosphatidic |
| chr21_-_14762944 | 0.00 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr10_+_4807913 | 0.00 |
ENSDART00000186099
|
palm2
|
paralemmin 2 |
| chr2_-_43151358 | 0.00 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr3_-_30434016 | 0.00 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr22_-_26251563 | 0.00 |
ENSDART00000060888
ENSDART00000142821 |
ccdc130
|
coiled-coil domain containing 130 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfat5a+nfat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0061182 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.5 | GO:0042182 | ketone catabolic process(GO:0042182) anthranilate metabolic process(GO:0043420) |
| 0.1 | 0.4 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.6 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:1990564 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |