Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
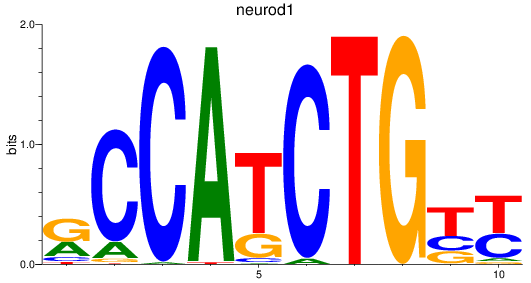
Results for neurod1
Z-value: 0.72
Transcription factors associated with neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
neurod1
|
ENSDARG00000019566 | neuronal differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| neurod1 | dr11_v1_chr9_-_44295071_44295071 | -0.66 | 3.1e-03 | Click! |
Activity profile of neurod1 motif
Sorted Z-values of neurod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10177442 | 2.03 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr1_+_218524 | 1.93 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr17_-_51829310 | 1.47 |
ENSDART00000154544
|
numb
|
numb homolog (Drosophila) |
| chr6_+_33076839 | 1.42 |
ENSDART00000073755
ENSDART00000122242 |
pomgnt1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr7_+_55950229 | 1.42 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr2_-_17114852 | 1.33 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr16_+_29509133 | 1.28 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr11_-_2270069 | 1.24 |
ENSDART00000189005
|
znf740a
|
zinc finger protein 740a |
| chr1_+_21937201 | 1.24 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr5_+_25762271 | 1.23 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr16_-_51151993 | 1.22 |
ENSDART00000156255
|
ago1
|
argonaute RISC catalytic component 1 |
| chr9_-_1200187 | 1.22 |
ENSDART00000158760
|
ino80da
|
INO80 complex subunit Da |
| chr3_-_30885250 | 1.21 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr5_+_36661058 | 1.18 |
ENSDART00000125653
|
capns1a
|
calpain, small subunit 1 a |
| chr2_-_17115256 | 1.14 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr11_-_25257045 | 1.12 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr2_-_49031303 | 1.11 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr3_-_61494840 | 1.10 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr3_-_29962345 | 1.10 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr11_-_25257595 | 1.10 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr4_-_20081621 | 1.08 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr1_+_12301913 | 1.07 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr5_+_37406358 | 1.06 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr13_+_22719789 | 1.04 |
ENSDART00000057672
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr11_-_43226255 | 1.03 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr1_-_34450784 | 1.03 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr17_+_12075805 | 1.02 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr15_-_26931541 | 1.02 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr1_-_34450622 | 1.01 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr11_-_35171162 | 1.01 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr3_-_23574622 | 1.00 |
ENSDART00000176012
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr18_-_39787040 | 1.00 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr1_-_55118745 | 0.99 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr20_+_405811 | 0.99 |
ENSDART00000149311
|
gpr63
|
G protein-coupled receptor 63 |
| chr1_-_9527200 | 0.98 |
ENSDART00000110790
|
si:ch73-12o23.1
|
si:ch73-12o23.1 |
| chr5_+_3891485 | 0.96 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr19_+_43669122 | 0.96 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr9_+_48123002 | 0.96 |
ENSDART00000099794
ENSDART00000169733 |
klhl23
|
kelch-like family member 23 |
| chr2_-_37478418 | 0.95 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr13_-_12667220 | 0.94 |
ENSDART00000079594
|
fam241a
|
family with sequence similarity 241 member A |
| chr21_-_18275226 | 0.94 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr19_-_868187 | 0.94 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr8_+_12118097 | 0.93 |
ENSDART00000081819
|
endog
|
endonuclease G |
| chr6_-_26225814 | 0.91 |
ENSDART00000089121
|
hs2st1b
|
heparan sulfate 2-O-sulfotransferase 1b |
| chr19_+_43684376 | 0.91 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr24_-_34335265 | 0.91 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr5_-_19932621 | 0.91 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr2_-_32237916 | 0.89 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr9_+_22634073 | 0.88 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr8_-_18613948 | 0.87 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr6_-_4214297 | 0.85 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr8_-_23776399 | 0.84 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr14_+_21722235 | 0.84 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr1_+_12302073 | 0.83 |
ENSDART00000164045
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr20_+_29209767 | 0.83 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr2_-_43545342 | 0.81 |
ENSDART00000179796
|
CABZ01058721.1
|
|
| chr10_-_15854743 | 0.81 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr18_+_3634652 | 0.80 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr8_-_49935358 | 0.79 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr20_+_29209926 | 0.78 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr17_+_1360192 | 0.78 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr9_+_25096500 | 0.77 |
ENSDART00000135074
ENSDART00000180436 ENSDART00000108629 |
lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr6_-_3992942 | 0.77 |
ENSDART00000182328
|
unc50
|
unc-50 homolog (C. elegans) |
| chr6_-_39700965 | 0.77 |
ENSDART00000156645
|
espl1
|
extra spindle pole bodies like 1, separase |
| chr7_+_34794829 | 0.76 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr5_+_6670945 | 0.76 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr20_+_29209615 | 0.76 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr24_+_19542323 | 0.74 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr8_-_19451487 | 0.74 |
ENSDART00000184037
|
zgc:92140
|
zgc:92140 |
| chr16_-_34212912 | 0.74 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr22_-_18241390 | 0.73 |
ENSDART00000083879
|
tmem161a
|
transmembrane protein 161A |
| chr18_+_45550783 | 0.71 |
ENSDART00000138075
|
kifc3
|
kinesin family member C3 |
| chr15_-_20745513 | 0.71 |
ENSDART00000187438
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr7_+_26844261 | 0.71 |
ENSDART00000079165
|
ext2
|
exostosin glycosyltransferase 2 |
| chr20_-_44576949 | 0.70 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr23_-_31647793 | 0.70 |
ENSDART00000145621
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr24_+_19518303 | 0.70 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr23_-_18057270 | 0.70 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr16_-_20855730 | 0.69 |
ENSDART00000188905
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr20_+_20731633 | 0.69 |
ENSDART00000191952
ENSDART00000165224 |
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr20_+_14968031 | 0.68 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr1_-_51710225 | 0.68 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr9_+_22929675 | 0.67 |
ENSDART00000061299
|
tsn
|
translin |
| chr24_+_19518570 | 0.67 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr3_-_54500354 | 0.67 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr8_+_20488322 | 0.66 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr23_-_18567088 | 0.66 |
ENSDART00000192371
|
sephs2
|
selenophosphate synthetase 2 |
| chr16_-_47408135 | 0.66 |
ENSDART00000150132
|
sept7b
|
septin 7b |
| chr16_+_35905031 | 0.66 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr2_+_35732652 | 0.66 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr5_+_62611400 | 0.66 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr3_+_33745014 | 0.65 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr2_+_47471647 | 0.65 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr11_-_44999858 | 0.65 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr19_-_32888758 | 0.65 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr19_-_43639331 | 0.64 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr15_-_28904371 | 0.63 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr2_+_37245382 | 0.63 |
ENSDART00000004626
|
sec62
|
SEC62 homolog, preprotein translocation factor |
| chr22_-_21897203 | 0.62 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr2_+_47471943 | 0.61 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr20_+_23498255 | 0.61 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr20_-_53963515 | 0.61 |
ENSDART00000110252
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr5_-_13315726 | 0.61 |
ENSDART00000143364
|
sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr21_-_43666420 | 0.60 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr20_+_52529044 | 0.60 |
ENSDART00000158230
ENSDART00000002787 |
pycr3
|
pyrroline-5-carboxylate reductase 3 |
| chr24_+_14801844 | 0.59 |
ENSDART00000141620
|
pi15a
|
peptidase inhibitor 15a |
| chr3_+_34988670 | 0.59 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr6_-_37749711 | 0.59 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr1_+_6646529 | 0.59 |
ENSDART00000144641
ENSDART00000103701 ENSDART00000138919 |
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr10_-_14943281 | 0.59 |
ENSDART00000143608
|
smad2
|
SMAD family member 2 |
| chr20_+_21268795 | 0.58 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr16_+_25171832 | 0.58 |
ENSDART00000156416
|
wu:fe05a04
|
wu:fe05a04 |
| chr20_-_18794789 | 0.58 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr9_+_48123224 | 0.58 |
ENSDART00000141610
|
klhl23
|
kelch-like family member 23 |
| chr17_+_32360673 | 0.57 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr19_-_12212692 | 0.57 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr23_-_18057851 | 0.56 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr7_-_29356084 | 0.56 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr25_-_18948816 | 0.56 |
ENSDART00000091549
|
nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr19_-_46037835 | 0.56 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr1_+_19538299 | 0.55 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr6_-_1187565 | 0.55 |
ENSDART00000191756
|
txnrd3
|
thioredoxin reductase 3 |
| chr20_-_3319642 | 0.55 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr25_-_12412704 | 0.55 |
ENSDART00000168275
|
det1
|
DET1, COP1 ubiquitin ligase partner |
| chr12_+_32292564 | 0.55 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr17_-_19466319 | 0.55 |
ENSDART00000170429
|
dicer1
|
dicer 1, ribonuclease type III |
| chr4_-_4592287 | 0.55 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr3_+_32651697 | 0.54 |
ENSDART00000055338
|
thoc6
|
THO complex 6 |
| chr23_-_19831739 | 0.54 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr22_+_22437561 | 0.54 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr18_+_12147971 | 0.53 |
ENSDART00000162067
ENSDART00000168386 |
fgd4a
|
FYVE, RhoGEF and PH domain containing 4a |
| chr2_-_31833347 | 0.53 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr7_+_6941583 | 0.53 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr22_-_18544701 | 0.53 |
ENSDART00000132156
|
cirbpb
|
cold inducible RNA binding protein b |
| chr16_+_35536075 | 0.53 |
ENSDART00000183618
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr5_+_42259002 | 0.52 |
ENSDART00000083778
|
eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr11_-_28911172 | 0.52 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr12_-_33659328 | 0.52 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr5_-_35252761 | 0.52 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr18_+_15778110 | 0.51 |
ENSDART00000014188
|
ube2na
|
ubiquitin-conjugating enzyme E2Na |
| chr9_+_35017702 | 0.51 |
ENSDART00000193640
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr11_+_6902946 | 0.51 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr10_+_6340478 | 0.50 |
ENSDART00000163182
ENSDART00000163722 |
tpm2
|
tropomyosin 2 (beta) |
| chr3_+_16663373 | 0.50 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr18_-_21047007 | 0.49 |
ENSDART00000162702
|
igf1ra
|
insulin-like growth factor 1a receptor |
| chr15_+_34963316 | 0.49 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr19_-_24136233 | 0.49 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr21_+_39100289 | 0.48 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr22_-_18240968 | 0.48 |
ENSDART00000027605
|
tmem161a
|
transmembrane protein 161A |
| chr13_-_8785610 | 0.48 |
ENSDART00000021083
|
calm2b
|
calmodulin 2b, (phosphorylase kinase, delta) |
| chr12_-_34852342 | 0.48 |
ENSDART00000152968
|
si:dkey-21c1.1
|
si:dkey-21c1.1 |
| chr6_-_1187749 | 0.48 |
ENSDART00000172544
|
txnrd3
|
thioredoxin reductase 3 |
| chr23_+_22873415 | 0.48 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr20_-_43741159 | 0.47 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr23_-_16980213 | 0.47 |
ENSDART00000046889
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr12_-_33459579 | 0.47 |
ENSDART00000105562
|
zgc:91940
|
zgc:91940 |
| chr17_+_28624321 | 0.47 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr23_-_18057553 | 0.47 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr24_+_35387517 | 0.47 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr5_+_60919378 | 0.46 |
ENSDART00000184915
|
doc2b
|
double C2-like domains, beta |
| chr7_-_59210882 | 0.46 |
ENSDART00000170330
ENSDART00000158996 |
nagk
|
N-acetylglucosamine kinase |
| chr12_-_22238004 | 0.46 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr21_-_41065369 | 0.46 |
ENSDART00000143749
|
larsb
|
leucyl-tRNA synthetase b |
| chr23_+_25172682 | 0.46 |
ENSDART00000191197
ENSDART00000183497 |
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr2_+_31308587 | 0.46 |
ENSDART00000027090
|
clul1
|
clusterin-like 1 (retinal) |
| chr10_-_14920989 | 0.45 |
ENSDART00000184617
|
smad2
|
SMAD family member 2 |
| chr21_+_11521163 | 0.45 |
ENSDART00000139267
|
zgc:114104
|
zgc:114104 |
| chr11_+_18873619 | 0.45 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr13_-_6250317 | 0.45 |
ENSDART00000180416
|
tuba4l
|
tubulin, alpha 4 like |
| chr5_-_40190949 | 0.45 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr15_+_29472065 | 0.44 |
ENSDART00000154343
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr9_-_39624173 | 0.44 |
ENSDART00000180106
ENSDART00000126766 |
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr12_+_21525496 | 0.44 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr21_+_6212844 | 0.44 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr24_-_26995164 | 0.44 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr5_-_51998708 | 0.43 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr2_-_32574944 | 0.43 |
ENSDART00000056642
|
tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr25_-_37258653 | 0.43 |
ENSDART00000131076
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr5_+_58455488 | 0.43 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr5_-_69923241 | 0.43 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr13_+_50778187 | 0.43 |
ENSDART00000159597
|
dntt
|
deoxynucleotidyltransferase, terminal |
| chr16_-_42066523 | 0.42 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr8_-_49207319 | 0.42 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr13_-_6252498 | 0.42 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr18_-_42313798 | 0.42 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr3_-_21061931 | 0.42 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr15_-_26568278 | 0.42 |
ENSDART00000182609
|
wdr81
|
WD repeat domain 81 |
| chr7_-_54217547 | 0.42 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr10_-_4961923 | 0.42 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr23_-_21758253 | 0.42 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr11_-_6880725 | 0.41 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr6_+_30533504 | 0.41 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr7_+_22399669 | 0.41 |
ENSDART00000144375
|
fgf11a
|
fibroblast growth factor 11a |
| chr6_-_51771634 | 0.41 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr21_+_8427059 | 0.41 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr19_-_42588510 | 0.40 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr5_-_30145939 | 0.39 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr12_+_19281189 | 0.38 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr10_-_6867282 | 0.38 |
ENSDART00000144001
ENSDART00000109744 |
ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr24_+_32472155 | 0.38 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
Network of associatons between targets according to the STRING database.
First level regulatory network of neurod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 1.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 1.0 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.3 | 0.9 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 1.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.3 | 2.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.3 | 0.8 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.2 | 1.5 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 2.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 0.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 1.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 0.8 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 1.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.5 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 2.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.7 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.9 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 1.9 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.3 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.7 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.4 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.4 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.4 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.4 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.6 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.4 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.8 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.4 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.7 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.9 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.2 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.4 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 1.0 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 2.0 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.8 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0072506 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.2 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.2 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.0 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 1.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.6 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 2.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 2.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 3.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.1 | GO:0098791 | Golgi stack(GO:0005795) Golgi subcompartment(GO:0098791) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.6 | 2.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.5 | 2.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 1.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 0.9 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.3 | 1.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.4 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.7 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.5 | GO:0070883 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.1 | 2.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.0 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.6 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.4 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.4 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.7 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 1.8 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.2 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.3 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.7 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.9 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 2.2 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.0 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |