Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
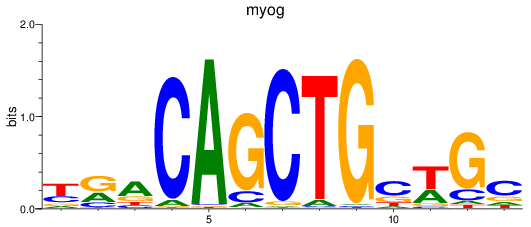
Results for myog
Z-value: 1.06
Transcription factors associated with myog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myog
|
ENSDARG00000009438 | myogenin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myog | dr11_v1_chr11_-_22605981_22605981 | -0.92 | 6.9e-08 | Click! |
Activity profile of myog motif
Sorted Z-values of myog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_14109348 | 3.59 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr7_-_20582842 | 2.80 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr2_-_11027258 | 2.79 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr15_+_25489406 | 2.72 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr20_+_34502606 | 2.29 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr21_-_19919020 | 2.26 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr6_+_153146 | 2.13 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr2_+_6253246 | 2.12 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr2_-_17115256 | 2.12 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_17114852 | 1.90 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr7_-_48263516 | 1.87 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr17_+_1360192 | 1.87 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr16_+_39159752 | 1.77 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr13_+_8840772 | 1.75 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr18_+_15271993 | 1.75 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr12_-_13729263 | 1.73 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr20_-_29498178 | 1.73 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_+_26212621 | 1.70 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr3_+_7771420 | 1.68 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr19_+_14454306 | 1.68 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr23_+_553396 | 1.59 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr21_-_38717854 | 1.59 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
| chr10_-_44560165 | 1.52 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr1_+_59321629 | 1.50 |
ENSDART00000161981
|
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr12_-_25380028 | 1.48 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr8_-_18613948 | 1.46 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr9_+_21306902 | 1.45 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr6_+_21001264 | 1.45 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr23_-_22523303 | 1.45 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr13_+_2894536 | 1.43 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr23_+_384850 | 1.39 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr12_-_10512911 | 1.35 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr3_-_40836081 | 1.35 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr23_-_18057270 | 1.33 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr8_-_1838315 | 1.30 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr14_-_34605607 | 1.28 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr21_-_217589 | 1.26 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr13_-_32726178 | 1.25 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr7_+_34794829 | 1.25 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr22_-_10541372 | 1.24 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr19_-_30810328 | 1.23 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr23_+_44614056 | 1.23 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr17_-_51818659 | 1.22 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr14_-_25928899 | 1.22 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr12_-_10508952 | 1.22 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr8_-_1255321 | 1.19 |
ENSDART00000149605
|
cdc14b
|
cell division cycle 14B |
| chr9_-_39624173 | 1.18 |
ENSDART00000180106
ENSDART00000126766 |
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr12_+_5251647 | 1.17 |
ENSDART00000124097
|
plce1
|
phospholipase C, epsilon 1 |
| chr19_-_47571456 | 1.17 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr1_+_27690 | 1.16 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr5_-_32336613 | 1.14 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_-_49854432 | 1.14 |
ENSDART00000180731
|
blvra
|
biliverdin reductase A |
| chr6_+_59176470 | 1.13 |
ENSDART00000161720
|
shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr21_+_4204860 | 1.12 |
ENSDART00000146541
|
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr8_+_26410539 | 1.11 |
ENSDART00000168780
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr21_+_33249478 | 1.09 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr5_-_20135679 | 1.07 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr25_+_3294150 | 1.07 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr6_+_40952031 | 1.06 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_+_51795667 | 1.06 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr6_-_14038804 | 1.05 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr11_+_45110865 | 1.04 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr20_+_23625387 | 1.04 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr5_+_37744625 | 1.04 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr25_+_17689565 | 1.03 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr5_+_31779911 | 1.03 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr20_+_1316495 | 1.02 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr16_+_29025292 | 0.98 |
ENSDART00000148865
ENSDART00000150184 |
gpatch4
|
G patch domain containing 4 |
| chr6_+_59991076 | 0.98 |
ENSDART00000163575
|
CABZ01100888.1
|
|
| chr13_+_11439486 | 0.97 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr20_-_44576949 | 0.97 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr22_-_21897203 | 0.97 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr23_+_45845159 | 0.96 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr20_+_329032 | 0.96 |
ENSDART00000036635
|
fynb
|
FYN proto-oncogene, Src family tyrosine kinase b |
| chr8_-_4573545 | 0.94 |
ENSDART00000127153
|
dhx37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr25_-_32869794 | 0.93 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr16_+_27614989 | 0.92 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr5_+_1515938 | 0.92 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr1_+_19764995 | 0.91 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr8_+_26410197 | 0.91 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr12_+_33320884 | 0.90 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr24_-_17389263 | 0.88 |
ENSDART00000122757
|
cul1b
|
cullin 1b |
| chr10_+_21444654 | 0.88 |
ENSDART00000140113
ENSDART00000184386 ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr16_+_14010242 | 0.88 |
ENSDART00000059928
|
fdps
|
farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr6_-_11759860 | 0.87 |
ENSDART00000151296
|
si:ch211-162i14.1
|
si:ch211-162i14.1 |
| chr9_-_32300783 | 0.87 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr9_-_32300611 | 0.86 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr23_-_10786400 | 0.86 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr23_-_18668836 | 0.86 |
ENSDART00000138792
ENSDART00000051182 |
arhgap4b
|
Rho GTPase activating protein 4b |
| chr14_-_46832825 | 0.85 |
ENSDART00000149571
|
ldb2a
|
LIM domain binding 2a |
| chr19_+_32257472 | 0.85 |
ENSDART00000186471
|
atxn1a
|
ataxin 1a |
| chr14_-_34605804 | 0.85 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr18_+_38191346 | 0.84 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr22_-_10541712 | 0.84 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr23_-_31266586 | 0.84 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr15_-_10455438 | 0.83 |
ENSDART00000158958
ENSDART00000192971 ENSDART00000162133 ENSDART00000185071 ENSDART00000192444 ENSDART00000165668 ENSDART00000175825 |
tenm4
|
teneurin transmembrane protein 4 |
| chr18_+_30441740 | 0.83 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr25_+_16214854 | 0.83 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr15_-_31109760 | 0.81 |
ENSDART00000154254
|
ksr1b
|
kinase suppressor of ras 1b |
| chr7_-_41726657 | 0.81 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr5_-_13315726 | 0.81 |
ENSDART00000143364
|
sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr16_+_33953644 | 0.80 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr14_+_21828993 | 0.80 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr23_+_45845423 | 0.79 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr23_-_31810222 | 0.78 |
ENSDART00000134319
ENSDART00000139076 |
hbs1l
|
HBS1-like translational GTPase |
| chr3_-_57762247 | 0.77 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr19_-_47570672 | 0.77 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr11_-_18015534 | 0.76 |
ENSDART00000181953
|
qrich1
|
glutamine-rich 1 |
| chr11_-_45171139 | 0.76 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr19_+_15441022 | 0.76 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr23_+_43718865 | 0.76 |
ENSDART00000175192
|
anapc10
|
anaphase promoting complex subunit 10 |
| chr4_-_4834347 | 0.75 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr2_-_37312927 | 0.75 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr5_+_17780475 | 0.74 |
ENSDART00000110783
ENSDART00000115227 |
chfr
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr25_+_34862225 | 0.74 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr18_-_20444296 | 0.74 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr25_-_17587785 | 0.73 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr8_-_1219815 | 0.73 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr1_+_49266886 | 0.73 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr13_-_36911118 | 0.73 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr10_+_15454745 | 0.72 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr5_-_24812715 | 0.72 |
ENSDART00000140042
ENSDART00000137128 ENSDART00000182415 ENSDART00000190898 |
ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr16_-_9869056 | 0.72 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr4_-_16658514 | 0.72 |
ENSDART00000133837
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr1_+_54124209 | 0.72 |
ENSDART00000187730
|
LO017722.1
|
|
| chr13_-_12021566 | 0.71 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr1_+_8508753 | 0.71 |
ENSDART00000091683
|
alkbh5
|
alkB homolog 5, RNA demethylase |
| chr16_-_42750295 | 0.70 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr13_-_13030851 | 0.70 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr7_-_45993416 | 0.70 |
ENSDART00000157602
ENSDART00000166983 |
zgc:101715
|
zgc:101715 |
| chr7_-_38570299 | 0.70 |
ENSDART00000143815
|
celf1
|
cugbp, Elav-like family member 1 |
| chr7_-_73856073 | 0.70 |
ENSDART00000181793
|
FP236812.2
|
|
| chr5_-_29195063 | 0.69 |
ENSDART00000109926
|
man1b1b
|
mannosidase, alpha, class 1B, member 1b |
| chr4_+_9055488 | 0.69 |
ENSDART00000155385
|
slc41a2b
|
solute carrier family 41 (magnesium transporter), member 2b |
| chr14_+_20156477 | 0.69 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr2_-_57918314 | 0.68 |
ENSDART00000138265
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr2_-_31833347 | 0.67 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr5_-_23574234 | 0.66 |
ENSDART00000002453
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr1_-_8566567 | 0.66 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr16_+_8139515 | 0.64 |
ENSDART00000193303
ENSDART00000139432 |
pomgnt2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr5_+_36781732 | 0.64 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr23_-_18057851 | 0.64 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr5_+_31811662 | 0.64 |
ENSDART00000023463
|
uap1l1
|
UDP-N-acetylglucosamine pyrophosphorylase 1, like 1 |
| chr23_-_1583193 | 0.64 |
ENSDART00000143841
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr9_+_21795917 | 0.63 |
ENSDART00000169069
|
rev1
|
REV1, polymerase (DNA directed) |
| chr5_-_18911114 | 0.63 |
ENSDART00000014434
|
bri3bp
|
bri3 binding protein |
| chr25_-_20666754 | 0.63 |
ENSDART00000158418
|
csk
|
C-terminal Src kinase |
| chr16_-_19568795 | 0.62 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr23_+_9508538 | 0.62 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr12_-_6063328 | 0.62 |
ENSDART00000002583
|
aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr13_-_8692860 | 0.62 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr12_+_5530247 | 0.61 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr9_+_46745348 | 0.61 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr23_-_19831739 | 0.61 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr8_+_104114 | 0.61 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr13_+_51710725 | 0.60 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr2_+_29995590 | 0.60 |
ENSDART00000151906
|
rbm33b
|
RNA binding motif protein 33b |
| chr5_+_8964926 | 0.60 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr18_+_6641542 | 0.60 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr20_-_53176675 | 0.59 |
ENSDART00000184013
|
CABZ01115113.1
|
|
| chr9_-_12885201 | 0.59 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr2_+_36620011 | 0.59 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr17_+_24064014 | 0.58 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr13_-_18011168 | 0.58 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr25_-_20666328 | 0.57 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr24_+_26337623 | 0.57 |
ENSDART00000145637
|
mynn
|
myoneurin |
| chr19_+_15440841 | 0.57 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr5_+_58455488 | 0.57 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr10_-_25328814 | 0.57 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr14_-_31694274 | 0.56 |
ENSDART00000173353
|
map7d3
|
MAP7 domain containing 3 |
| chr11_+_39107131 | 0.55 |
ENSDART00000105140
|
zgc:112255
|
zgc:112255 |
| chr6_+_46309795 | 0.55 |
ENSDART00000154817
|
si:dkeyp-67f1.1
|
si:dkeyp-67f1.1 |
| chr15_+_24746444 | 0.55 |
ENSDART00000032306
|
ywhae1
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 |
| chr5_+_4016271 | 0.55 |
ENSDART00000113627
ENSDART00000105832 ENSDART00000121415 |
ggnbp2
|
gametogenetin binding protein 2 |
| chr2_-_7185460 | 0.54 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr14_-_28534563 | 0.54 |
ENSDART00000054088
|
zgc:113364
|
zgc:113364 |
| chr23_-_27505825 | 0.54 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr24_-_31090948 | 0.53 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr23_-_18057553 | 0.53 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr15_-_47115787 | 0.53 |
ENSDART00000192601
|
CABZ01079081.1
|
|
| chr4_-_25257451 | 0.53 |
ENSDART00000142819
|
tmem110l
|
transmembrane protein 110, like |
| chr20_-_15161669 | 0.53 |
ENSDART00000080333
ENSDART00000063882 |
plpp6
|
phospholipid phosphatase 6 |
| chr25_-_25619550 | 0.52 |
ENSDART00000150400
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr12_-_10476448 | 0.52 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr23_+_41831224 | 0.52 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr5_+_36768674 | 0.52 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr3_+_46559639 | 0.51 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr13_+_27770424 | 0.51 |
ENSDART00000159281
|
asrgl1
|
asparaginase like 1 |
| chr1_+_45925365 | 0.51 |
ENSDART00000144245
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr18_+_5454341 | 0.51 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr13_-_11743973 | 0.51 |
ENSDART00000190641
ENSDART00000079310 |
manba
|
mannosidase, beta A, lysosomal |
| chr8_-_46386024 | 0.50 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr15_+_34963316 | 0.50 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr8_+_387622 | 0.50 |
ENSDART00000167361
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr22_+_18929412 | 0.50 |
ENSDART00000161598
ENSDART00000166650 ENSDART00000015951 ENSDART00000105392 ENSDART00000131131 |
bsg
|
basigin |
| chr8_+_14959587 | 0.50 |
ENSDART00000138548
ENSDART00000041697 |
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr13_+_24263049 | 0.49 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr15_-_4568154 | 0.49 |
ENSDART00000155254
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr14_+_30774032 | 0.49 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr9_-_41025062 | 0.49 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
Network of associatons between targets according to the STRING database.
First level regulatory network of myog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 1.7 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.4 | 1.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 1.7 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.4 | 1.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.4 | 1.9 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.3 | 4.0 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.3 | 3.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 1.1 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.2 | 1.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.2 | 0.7 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 0.6 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.8 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.2 | 0.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 0.9 | GO:1990592 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.7 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.7 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.2 | 2.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.9 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.7 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.5 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 2.1 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 2.8 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 1.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.6 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.5 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.1 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 0.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.5 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.1 | 1.7 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 3.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.3 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 1.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 1.0 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.4 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.5 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.5 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.7 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.1 | 0.3 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 1.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.3 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.3 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.4 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 1.9 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.4 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.0 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.2 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 1.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.9 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 2.2 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.6 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 1.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.0 | 1.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.5 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 2.6 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 1.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0008645 | hexose transport(GO:0008645) glucose transport(GO:0015758) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.4 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.0 | 1.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.2 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 4.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.7 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 3.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.7 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 5.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.9 | GO:0032153 | cell division site(GO:0032153) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.5 | 3.7 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.5 | 1.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 1.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.4 | 1.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 0.9 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 2.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.6 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.7 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.2 | 1.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 2.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.8 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.6 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.5 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.2 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.6 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 2.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 1.3 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.7 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 2.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 3.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 0.9 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 3.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |