Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
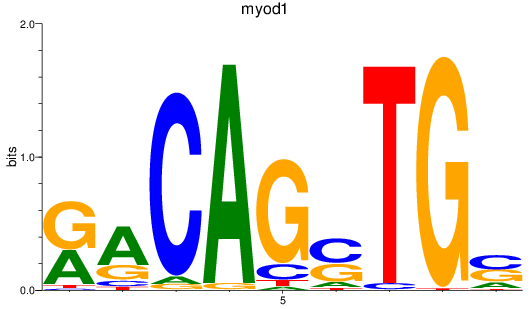
Results for myod1
Z-value: 0.33
Transcription factors associated with myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myod1
|
ENSDARG00000030110 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myod1 | dr11_v1_chr25_-_31423493_31423493 | 0.37 | 1.3e-01 | Click! |
Activity profile of myod1 motif
Sorted Z-values of myod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_42467867 | 0.49 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr25_-_19585010 | 0.46 |
ENSDART00000021340
|
sycp3
|
synaptonemal complex protein 3 |
| chr3_-_31783737 | 0.41 |
ENSDART00000090809
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr3_-_31784082 | 0.39 |
ENSDART00000134201
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr22_+_18187857 | 0.38 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr8_-_51349429 | 0.38 |
ENSDART00000134043
ENSDART00000098282 |
si:ch211-198o12.4
|
si:ch211-198o12.4 |
| chr25_-_19584735 | 0.37 |
ENSDART00000137930
|
sycp3
|
synaptonemal complex protein 3 |
| chr24_+_17269849 | 0.37 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr5_+_42402536 | 0.36 |
ENSDART00000186754
|
BX548073.11
|
|
| chr25_-_16755340 | 0.33 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr5_-_46273938 | 0.33 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr10_-_35103208 | 0.33 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr6_+_7249531 | 0.32 |
ENSDART00000125912
ENSDART00000083424 ENSDART00000049695 ENSDART00000136088 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr23_+_19977120 | 0.31 |
ENSDART00000089342
|
cfap126
|
cilia and flagella associated protein 126 |
| chr25_-_4482449 | 0.31 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr12_-_17810543 | 0.31 |
ENSDART00000090484
|
tecpr1a
|
tectonin beta-propeller repeat containing 1a |
| chr16_+_32136550 | 0.30 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr22_-_29906764 | 0.30 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr7_+_22616212 | 0.30 |
ENSDART00000052844
|
cldn7a
|
claudin 7a |
| chr22_+_11756040 | 0.30 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr8_-_23194408 | 0.30 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr6_-_6346557 | 0.29 |
ENSDART00000115018
|
ccdc88aa
|
coiled-coil domain containing 88Aa |
| chr3_+_17910569 | 0.29 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr22_+_18188045 | 0.28 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr2_+_27491066 | 0.28 |
ENSDART00000078305
|
zswim5
|
zinc finger, SWIM-type containing 5 |
| chr13_-_5978433 | 0.27 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr22_+_12798569 | 0.27 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr1_-_34685007 | 0.26 |
ENSDART00000157471
|
klf5a
|
Kruppel-like factor 5a |
| chr9_-_24970018 | 0.26 |
ENSDART00000026924
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr24_+_37902687 | 0.26 |
ENSDART00000042273
|
ccdc78
|
coiled-coil domain containing 78 |
| chr19_-_23892243 | 0.26 |
ENSDART00000132384
|
si:dkey-222b8.4
|
si:dkey-222b8.4 |
| chr8_+_21376290 | 0.25 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr3_-_29962345 | 0.25 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr2_-_42492201 | 0.24 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr7_+_39402864 | 0.24 |
ENSDART00000025852
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr3_-_61592417 | 0.24 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr10_+_42358426 | 0.23 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr5_-_61787969 | 0.23 |
ENSDART00000112744
|
gas2l2
|
growth arrest specific 2 like 2 |
| chr25_+_3549401 | 0.23 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr17_+_41992054 | 0.23 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr12_-_33646010 | 0.23 |
ENSDART00000111259
|
tmem94
|
transmembrane protein 94 |
| chr11_-_6001845 | 0.23 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr8_-_11229523 | 0.23 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr10_-_625441 | 0.23 |
ENSDART00000171171
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr1_-_52498146 | 0.22 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_-_29146624 | 0.22 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr5_-_52569014 | 0.22 |
ENSDART00000165616
|
mamdc2a
|
MAM domain containing 2a |
| chr24_-_35707552 | 0.21 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr18_+_402048 | 0.21 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr8_+_50942240 | 0.21 |
ENSDART00000124748
|
b2ml
|
beta-2-microglobulin, like |
| chr3_-_19200571 | 0.21 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr3_-_26204867 | 0.21 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr5_-_3991655 | 0.21 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr10_-_690072 | 0.20 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr22_-_10110959 | 0.20 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr19_-_8877469 | 0.20 |
ENSDART00000193951
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr23_-_24682244 | 0.20 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr23_-_29003864 | 0.19 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr16_-_42894628 | 0.19 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr10_+_6884123 | 0.19 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr17_+_14425219 | 0.19 |
ENSDART00000153994
|
ccdc175
|
coiled-coil domain containing 175 |
| chr19_-_7115229 | 0.19 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr23_+_23658474 | 0.19 |
ENSDART00000162838
|
agrn
|
agrin |
| chr20_-_21672970 | 0.19 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr4_-_4592287 | 0.19 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr5_-_69212184 | 0.19 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr22_+_465269 | 0.19 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr13_+_30903816 | 0.19 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr25_-_14637660 | 0.19 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr4_+_20177526 | 0.19 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr23_+_27789795 | 0.18 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr25_+_4541211 | 0.18 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr20_+_41549200 | 0.18 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr12_-_33659328 | 0.18 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr12_-_32013125 | 0.18 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr21_-_43126881 | 0.18 |
ENSDART00000174463
ENSDART00000160845 |
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr8_-_49935358 | 0.17 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr6_-_16393491 | 0.17 |
ENSDART00000179312
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr3_-_16227490 | 0.17 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr20_-_3319642 | 0.17 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr5_+_19867794 | 0.17 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr7_-_54217547 | 0.17 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr25_-_3644574 | 0.17 |
ENSDART00000188569
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr21_+_27382893 | 0.17 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr5_-_38384289 | 0.17 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr5_-_17601759 | 0.17 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr1_+_49352900 | 0.16 |
ENSDART00000008468
|
msx1b
|
muscle segment homeobox 1b |
| chr1_-_52497834 | 0.16 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_+_6652967 | 0.16 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr10_+_35257651 | 0.16 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr18_-_14337065 | 0.16 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr23_-_21758253 | 0.16 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr7_+_72460911 | 0.16 |
ENSDART00000160682
ENSDART00000168532 |
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr25_-_3549321 | 0.16 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr22_+_12366516 | 0.16 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr17_-_28097760 | 0.16 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr13_-_10672538 | 0.16 |
ENSDART00000132393
|
slc3a1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr14_-_25956804 | 0.15 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr13_-_7767548 | 0.15 |
ENSDART00000158720
ENSDART00000121952 |
h2afy2
|
H2A histone family, member Y2 |
| chr8_+_53269657 | 0.15 |
ENSDART00000184212
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr7_+_49715750 | 0.15 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr17_-_38887424 | 0.15 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr18_-_14337450 | 0.15 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr14_-_36862745 | 0.15 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr21_+_17542473 | 0.15 |
ENSDART00000005750
ENSDART00000141326 |
stom
|
stomatin |
| chr9_+_33158191 | 0.15 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr6_-_12456077 | 0.15 |
ENSDART00000190107
|
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr19_-_5369486 | 0.15 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr22_+_22021936 | 0.15 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr4_-_75175407 | 0.15 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr12_-_20795867 | 0.15 |
ENSDART00000152835
ENSDART00000153424 |
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr19_-_35035857 | 0.15 |
ENSDART00000103253
|
bmp8a
|
bone morphogenetic protein 8a |
| chr21_-_41369370 | 0.15 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr20_+_23408970 | 0.15 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr7_+_24049776 | 0.15 |
ENSDART00000166559
|
efs
|
embryonal Fyn-associated substrate |
| chr8_-_48740953 | 0.14 |
ENSDART00000160805
|
pimr180
|
Pim proto-oncogene, serine/threonine kinase, related 180 |
| chr4_-_8043839 | 0.14 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr19_-_24555935 | 0.14 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr19_-_6134802 | 0.14 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr16_-_4610255 | 0.14 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr1_-_17650223 | 0.14 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr5_+_62611400 | 0.14 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr16_+_41060161 | 0.14 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr16_+_3004422 | 0.14 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr4_+_8670662 | 0.14 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
| chr10_+_25355308 | 0.14 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr21_+_6212844 | 0.14 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr4_-_8093753 | 0.14 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr4_-_18635005 | 0.14 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr7_-_67894046 | 0.14 |
ENSDART00000168964
|
si:ch73-315f9.2
|
si:ch73-315f9.2 |
| chr24_+_39034090 | 0.14 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr14_+_31651533 | 0.14 |
ENSDART00000172835
|
fhl1a
|
four and a half LIM domains 1a |
| chr7_+_56253914 | 0.14 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr3_+_37197686 | 0.14 |
ENSDART00000151144
|
fmnl1a
|
formin-like 1a |
| chr5_-_34185115 | 0.13 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr4_-_12978925 | 0.13 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr7_-_58269812 | 0.13 |
ENSDART00000050077
|
sdcbp
|
syndecan binding protein (syntenin) |
| chr13_-_7767044 | 0.13 |
ENSDART00000159453
|
h2afy2
|
H2A histone family, member Y2 |
| chr8_-_18225968 | 0.13 |
ENSDART00000135504
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr14_-_29905962 | 0.13 |
ENSDART00000142605
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr10_-_6867282 | 0.13 |
ENSDART00000144001
ENSDART00000109744 |
ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr14_-_17563773 | 0.13 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr22_+_11775269 | 0.13 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr13_-_27660955 | 0.13 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr13_-_31452516 | 0.13 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr24_+_10414028 | 0.13 |
ENSDART00000193257
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr14_+_24840669 | 0.13 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr16_-_35585374 | 0.13 |
ENSDART00000180208
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr8_-_8698607 | 0.13 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr20_-_33497128 | 0.13 |
ENSDART00000101974
|
erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr8_-_1219815 | 0.13 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr2_+_6181383 | 0.13 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr8_-_48675152 | 0.13 |
ENSDART00000164544
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr17_+_23938283 | 0.13 |
ENSDART00000184391
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr1_-_45750391 | 0.12 |
ENSDART00000133979
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr11_-_21528056 | 0.12 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr20_+_47434709 | 0.12 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr17_-_15667198 | 0.12 |
ENSDART00000142972
ENSDART00000132571 ENSDART00000189936 |
manea
|
mannosidase, endo-alpha |
| chr22_-_18241390 | 0.12 |
ENSDART00000083879
|
tmem161a
|
transmembrane protein 161A |
| chr6_-_39051319 | 0.12 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr14_+_31657412 | 0.12 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr19_-_24555623 | 0.12 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr3_-_21061931 | 0.12 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr1_-_34685329 | 0.12 |
ENSDART00000125944
ENSDART00000008277 |
pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr10_-_1718395 | 0.12 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr2_-_29996036 | 0.12 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr9_-_105135 | 0.12 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr19_+_1673599 | 0.12 |
ENSDART00000163127
|
klhl7
|
kelch-like family member 7 |
| chr5_-_13818833 | 0.12 |
ENSDART00000192198
|
add2
|
adducin 2 (beta) |
| chr15_+_27328089 | 0.12 |
ENSDART00000079294
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr1_-_38709551 | 0.12 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr17_+_15041647 | 0.11 |
ENSDART00000108999
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr10_-_25069155 | 0.11 |
ENSDART00000078226
ENSDART00000181941 |
mtnr1bb
|
melatonin receptor 1Bb |
| chr11_+_13423776 | 0.11 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr14_+_46410766 | 0.11 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr2_+_49864219 | 0.11 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr19_+_31904836 | 0.11 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr9_+_41024973 | 0.11 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr17_+_23968214 | 0.11 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr13_-_7766758 | 0.11 |
ENSDART00000171831
|
h2afy2
|
H2A histone family, member Y2 |
| chr7_-_39378903 | 0.11 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr23_-_14918276 | 0.11 |
ENSDART00000179831
|
ndrg3b
|
ndrg family member 3b |
| chr22_+_17214389 | 0.11 |
ENSDART00000187083
ENSDART00000159468 ENSDART00000129109 ENSDART00000090107 |
nrd1b
|
nardilysin b (N-arginine dibasic convertase) |
| chr16_-_13789908 | 0.11 |
ENSDART00000138540
|
ttyh1
|
tweety family member 1 |
| chr16_-_29164379 | 0.11 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr14_+_49602 | 0.11 |
ENSDART00000035581
|
OTOP1
|
otopetrin 1 |
| chr9_-_7655243 | 0.11 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr5_-_60159116 | 0.11 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr14_-_30704075 | 0.11 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr8_+_45294767 | 0.11 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr24_+_744713 | 0.11 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr16_-_35579200 | 0.11 |
ENSDART00000162172
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr25_-_8602437 | 0.11 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr19_+_41551543 | 0.11 |
ENSDART00000112364
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr5_-_64431927 | 0.11 |
ENSDART00000158248
|
brd3b
|
bromodomain containing 3b |
| chr6_-_39764995 | 0.11 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr6_+_11989537 | 0.11 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr22_-_18240968 | 0.11 |
ENSDART00000027605
|
tmem161a
|
transmembrane protein 161A |
| chr6_+_29693492 | 0.11 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr19_-_10612774 | 0.11 |
ENSDART00000091898
|
si:dkey-211g8.6
|
si:dkey-211g8.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of myod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.2 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.3 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.3 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0090156 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.3 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.3 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.0 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |