Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
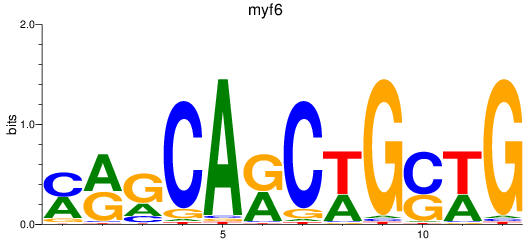
Results for myf6
Z-value: 0.85
Transcription factors associated with myf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myf6
|
ENSDARG00000029830 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myf6 | dr11_v1_chr4_+_21717793_21717793 | 0.53 | 2.4e-02 | Click! |
Activity profile of myf6 motif
Sorted Z-values of myf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_45319592 | 2.03 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr17_-_5769196 | 1.78 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr11_-_6001845 | 1.67 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr13_+_27951688 | 1.45 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr5_+_42381273 | 1.44 |
ENSDART00000142387
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr6_+_9241121 | 1.42 |
ENSDART00000064989
|
pimr70
|
Pim proto-oncogene, serine/threonine kinase, related 70 |
| chr9_-_105135 | 1.33 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr7_+_44715224 | 1.32 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr5_-_29671586 | 1.26 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr10_-_625441 | 1.26 |
ENSDART00000171171
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr6_-_9459440 | 1.23 |
ENSDART00000064991
|
pimr71
|
Pim proto-oncogene, serine/threonine kinase, related 71 |
| chr6_-_39313027 | 1.20 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr19_-_5332784 | 1.20 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr13_-_7767044 | 1.19 |
ENSDART00000159453
|
h2afy2
|
H2A histone family, member Y2 |
| chr21_+_198269 | 1.15 |
ENSDART00000193485
ENSDART00000163696 |
gsdf
|
gonadal somatic cell derived factor |
| chr17_-_39786222 | 1.11 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr5_-_71705191 | 1.10 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr22_-_3344613 | 1.08 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr21_-_18972206 | 1.07 |
ENSDART00000146743
|
pimr72
|
Pim proto-oncogene, serine/threonine kinase, related 72 |
| chr13_-_7766758 | 1.04 |
ENSDART00000171831
|
h2afy2
|
H2A histone family, member Y2 |
| chr1_-_625875 | 1.02 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr5_+_29671681 | 1.02 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr22_-_5099824 | 1.02 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr25_+_1035656 | 1.01 |
ENSDART00000179843
ENSDART00000168637 |
si:ch211-284k5.2
|
si:ch211-284k5.2 |
| chr23_-_45318760 | 1.01 |
ENSDART00000166883
|
ccdc171
|
coiled-coil domain containing 171 |
| chr8_-_50259448 | 0.98 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr10_-_690072 | 0.95 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr14_-_36862745 | 0.95 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr1_+_57311901 | 0.94 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr5_-_36948586 | 0.93 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr5_+_19867794 | 0.90 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr9_-_48407408 | 0.89 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr19_+_31904836 | 0.89 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr25_+_245018 | 0.88 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr5_+_42467867 | 0.87 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr2_-_7696287 | 0.86 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr5_-_19400166 | 0.85 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr23_-_12423778 | 0.83 |
ENSDART00000124091
|
wfdc2
|
WAP four-disulfide core domain 2 |
| chr21_-_3452683 | 0.83 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr3_-_22829710 | 0.82 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr20_-_9199721 | 0.82 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr4_+_69619 | 0.81 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
| chr17_-_7861219 | 0.79 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr4_+_90048 | 0.78 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr7_-_20756013 | 0.78 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr9_-_68934 | 0.77 |
ENSDART00000054594
ENSDART00000009389 |
il10rb
|
interleukin 10 receptor, beta |
| chr25_-_13188214 | 0.76 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr5_+_62723233 | 0.76 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr6_+_9893554 | 0.76 |
ENSDART00000064979
|
pimr74
|
Pim proto-oncogene, serine/threonine kinase, related 74 |
| chr12_+_18533198 | 0.75 |
ENSDART00000189729
|
meiob
|
meiosis specific with OB domains |
| chr1_-_52498146 | 0.74 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr2_+_27491066 | 0.74 |
ENSDART00000078305
|
zswim5
|
zinc finger, SWIM-type containing 5 |
| chr7_+_1467863 | 0.74 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr25_-_4482449 | 0.73 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr8_+_54188978 | 0.72 |
ENSDART00000175913
|
FO904970.1
|
|
| chr12_-_145076 | 0.72 |
ENSDART00000160926
|
dnah9
|
dynein, axonemal, heavy chain 9 |
| chr12_-_47793857 | 0.71 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr13_-_37619159 | 0.71 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr22_+_35472653 | 0.70 |
ENSDART00000076424
ENSDART00000187204 |
tctex1d2
|
Tctex1 domain containing 2 |
| chr5_+_42402536 | 0.69 |
ENSDART00000186754
|
BX548073.11
|
|
| chr4_+_61171310 | 0.69 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr13_-_438705 | 0.68 |
ENSDART00000082142
|
CU570800.1
|
|
| chr22_-_38607504 | 0.66 |
ENSDART00000164609
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr16_+_52512025 | 0.66 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr3_+_26145013 | 0.65 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr11_-_30636163 | 0.65 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr20_+_52774730 | 0.63 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr12_-_11570 | 0.63 |
ENSDART00000186179
|
shisa6
|
shisa family member 6 |
| chr18_+_41250938 | 0.63 |
ENSDART00000087566
|
trip12
|
thyroid hormone receptor interactor 12 |
| chr7_-_59256806 | 0.63 |
ENSDART00000167955
|
m1ap
|
meiosis 1 associated protein |
| chr8_+_26522013 | 0.62 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr10_-_44027391 | 0.62 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr19_+_31404686 | 0.60 |
ENSDART00000078459
|
pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr18_-_44285539 | 0.60 |
ENSDART00000137222
|
pimr179
|
Pim proto-oncogene, serine/threonine kinase, related 179 |
| chr25_+_37480285 | 0.60 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr4_-_64709908 | 0.60 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr22_+_24715282 | 0.60 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr6_-_9922266 | 0.59 |
ENSDART00000151549
|
pimr73
|
Pim proto-oncogene, serine/threonine kinase, related 73 |
| chr13_-_28674422 | 0.57 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr10_+_35265083 | 0.57 |
ENSDART00000048831
|
tmem120a
|
transmembrane protein 120A |
| chr1_-_59407322 | 0.57 |
ENSDART00000161661
|
si:ch211-188p14.5
|
si:ch211-188p14.5 |
| chr2_-_7696503 | 0.56 |
ENSDART00000169709
|
CABZ01055306.1
|
|
| chr8_+_53928472 | 0.56 |
ENSDART00000063242
|
MMP23B
|
matrix metallopeptidase 23B |
| chr1_-_52497834 | 0.55 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr14_+_39255437 | 0.54 |
ENSDART00000147443
|
diaph2
|
diaphanous-related formin 2 |
| chr10_+_41986685 | 0.54 |
ENSDART00000086165
ENSDART00000171983 |
setd1ba
|
SET domain containing 1B, a |
| chr13_-_37615029 | 0.54 |
ENSDART00000111199
|
si:dkey-188i13.6
|
si:dkey-188i13.6 |
| chr4_-_38033800 | 0.53 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr13_-_37180815 | 0.53 |
ENSDART00000139907
|
si:dkeyp-77c8.1
|
si:dkeyp-77c8.1 |
| chr20_+_34029820 | 0.53 |
ENSDART00000143901
ENSDART00000134305 |
prg4b
|
proteoglycan 4b |
| chr19_-_42391383 | 0.53 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr1_-_52128425 | 0.53 |
ENSDART00000149939
|
rad23aa
|
RAD23 homolog A, nucleotide excision repair protein a |
| chr17_+_41992054 | 0.53 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr10_+_439692 | 0.52 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr3_-_3496738 | 0.52 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr21_+_18353703 | 0.52 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr21_-_30545121 | 0.52 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr10_+_16592851 | 0.51 |
ENSDART00000187508
ENSDART00000101142 |
chsy3
|
chondroitin sulfate synthase 3 |
| chr2_-_57891504 | 0.51 |
ENSDART00000139948
|
zgc:92789
|
zgc:92789 |
| chr23_+_4288964 | 0.51 |
ENSDART00000138408
|
l3mbtl1
|
l(3)mbt-like 1 (Drosophila) |
| chr11_-_8208464 | 0.51 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr21_+_45496442 | 0.51 |
ENSDART00000164880
|
si:dkey-223p19.1
|
si:dkey-223p19.1 |
| chr5_-_72136548 | 0.51 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr16_-_54405976 | 0.50 |
ENSDART00000055395
|
osr2
|
odd-skipped related transciption factor 2 |
| chr18_+_5549672 | 0.50 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr1_-_18446161 | 0.50 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr2_+_13462305 | 0.50 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr13_+_30903816 | 0.50 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr21_+_45366229 | 0.49 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr2_-_2937225 | 0.49 |
ENSDART00000168132
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr14_-_36863432 | 0.49 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr12_+_47945664 | 0.48 |
ENSDART00000189824
ENSDART00000055743 ENSDART00000188936 |
CABZ01074994.1
|
|
| chr13_-_6218248 | 0.48 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr23_-_46201008 | 0.48 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr8_-_29719393 | 0.48 |
ENSDART00000077635
|
si:ch211-103n10.5
|
si:ch211-103n10.5 |
| chr23_-_42752550 | 0.48 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr1_+_52398205 | 0.48 |
ENSDART00000143225
|
si:ch211-217k17.9
|
si:ch211-217k17.9 |
| chr4_+_10017049 | 0.48 |
ENSDART00000144175
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr23_-_19558556 | 0.47 |
ENSDART00000134012
|
dnah12
|
dynein, axonemal, heavy chain 12 |
| chr18_+_37259800 | 0.47 |
ENSDART00000078053
ENSDART00000132577 |
tbcb
|
tubulin folding cofactor B |
| chr12_+_18532866 | 0.47 |
ENSDART00000152126
ENSDART00000152443 ENSDART00000089589 |
meiob
|
meiosis specific with OB domains |
| chr12_+_49135755 | 0.47 |
ENSDART00000153460
|
si:zfos-911d5.4
|
si:zfos-911d5.4 |
| chr25_-_518656 | 0.47 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr23_+_44644911 | 0.46 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr8_-_6877390 | 0.46 |
ENSDART00000170883
ENSDART00000005321 |
neflb
|
neurofilament, light polypeptide b |
| chr24_+_26997798 | 0.46 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr12_+_18782821 | 0.45 |
ENSDART00000152918
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr2_-_26563978 | 0.45 |
ENSDART00000150016
|
glis1a
|
GLIS family zinc finger 1a |
| chr4_-_9764767 | 0.45 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr14_+_31657412 | 0.45 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr22_+_20720808 | 0.45 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr9_+_22657221 | 0.45 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr19_+_63567 | 0.45 |
ENSDART00000165657
ENSDART00000165183 |
zhx2b
|
zinc fingers and homeoboxes 2b |
| chr24_-_35707552 | 0.45 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_56431298 | 0.45 |
ENSDART00000182544
|
ctxn1
|
cortexin 1 |
| chr8_+_52788997 | 0.44 |
ENSDART00000169198
|
adgrd2
|
adhesion G protein-coupled receptor D2 |
| chr1_-_36151377 | 0.44 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr9_-_296169 | 0.44 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr25_+_37209619 | 0.44 |
ENSDART00000112192
|
si:dkey-234i14.3
|
si:dkey-234i14.3 |
| chr20_+_48100261 | 0.44 |
ENSDART00000158604
|
xkr5a
|
XK related 5a |
| chr9_+_41024973 | 0.44 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr8_+_53344726 | 0.44 |
ENSDART00000184395
ENSDART00000170212 |
CU914536.1
|
|
| chr13_+_48482256 | 0.44 |
ENSDART00000053332
|
FOXN2 (1 of many)
|
forkhead box N2 |
| chr19_+_551963 | 0.43 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr17_+_32500387 | 0.43 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr5_-_67895656 | 0.43 |
ENSDART00000158917
|
abhd10a
|
abhydrolase domain containing 10a |
| chr4_-_8093753 | 0.43 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr13_-_19799824 | 0.43 |
ENSDART00000058032
|
fam204a
|
family with sequence similarity 204, member A |
| chr10_-_5020378 | 0.42 |
ENSDART00000191857
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr20_-_29787192 | 0.42 |
ENSDART00000125348
ENSDART00000048759 |
id2b
|
inhibitor of DNA binding 2b |
| chr5_+_2815021 | 0.42 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr25_-_3549321 | 0.42 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr5_+_43530388 | 0.42 |
ENSDART00000190254
ENSDART00000097618 ENSDART00000133006 |
si:dkey-40c11.2
|
si:dkey-40c11.2 |
| chr5_-_46273938 | 0.42 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr17_+_46739693 | 0.42 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr23_+_23658474 | 0.42 |
ENSDART00000162838
|
agrn
|
agrin |
| chr14_+_15191176 | 0.42 |
ENSDART00000183447
ENSDART00000193093 ENSDART00000169309 |
si:dkey-203a12.2
|
si:dkey-203a12.2 |
| chr23_-_19558263 | 0.41 |
ENSDART00000084477
ENSDART00000179748 ENSDART00000193127 |
dnah12
|
dynein, axonemal, heavy chain 12 |
| chr7_-_27037990 | 0.41 |
ENSDART00000173561
|
nucb2a
|
nucleobindin 2a |
| chr14_+_41345175 | 0.41 |
ENSDART00000086104
|
nox1
|
NADPH oxidase 1 |
| chr7_+_17816470 | 0.41 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr21_-_32467799 | 0.41 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr25_-_4525081 | 0.41 |
ENSDART00000184347
|
pidd1
|
p53-induced death domain protein 1 |
| chr12_-_17810543 | 0.41 |
ENSDART00000090484
|
tecpr1a
|
tectonin beta-propeller repeat containing 1a |
| chr2_-_58902477 | 0.41 |
ENSDART00000061638
ENSDART00000172822 |
mau2
|
MAU2 sister chromatid cohesion factor |
| chr14_-_33454595 | 0.40 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr2_-_183992 | 0.40 |
ENSDART00000126704
ENSDART00000191283 ENSDART00000034783 |
zgc:113518
|
zgc:113518 |
| chr10_+_10351685 | 0.40 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr19_-_46957968 | 0.40 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr1_-_46505310 | 0.40 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr19_+_48410195 | 0.40 |
ENSDART00000166877
|
si:ch1073-376c22.1
|
si:ch1073-376c22.1 |
| chr7_-_73848458 | 0.40 |
ENSDART00000041385
|
zgc:163061
|
zgc:163061 |
| chr7_+_34506937 | 0.39 |
ENSDART00000111303
|
rfx7a
|
regulatory factor X7a |
| chr4_+_12358822 | 0.39 |
ENSDART00000172557
ENSDART00000150627 |
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr3_+_1211242 | 0.39 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr14_-_46310549 | 0.39 |
ENSDART00000123832
|
crybb1l1
|
crystallin, beta B1, like 1 |
| chr17_+_23255365 | 0.39 |
ENSDART00000180277
|
AL935174.5
|
|
| chr20_+_591505 | 0.39 |
ENSDART00000046438
|
kcnk2b
|
potassium channel, subfamily K, member 2b |
| chr13_+_681628 | 0.39 |
ENSDART00000016604
|
abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr2_-_20599315 | 0.39 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr14_+_45732081 | 0.38 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr10_+_17592273 | 0.38 |
ENSDART00000141221
|
KCNN2
|
si:dkey-76p7.5 |
| chr15_-_19771981 | 0.38 |
ENSDART00000175502
ENSDART00000159475 |
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr6_+_11989537 | 0.38 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr7_+_19903924 | 0.38 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr25_+_3549401 | 0.38 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr17_-_6600899 | 0.38 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr3_-_56924654 | 0.38 |
ENSDART00000157038
|
hid1a
|
HID1 domain containing a |
| chr10_+_25355308 | 0.38 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr18_+_37272568 | 0.37 |
ENSDART00000132749
|
tmem123
|
transmembrane protein 123 |
| chr6_+_53429228 | 0.37 |
ENSDART00000165067
|
abhd14b
|
abhydrolase domain containing 14B |
| chr21_-_43040780 | 0.37 |
ENSDART00000187037
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr23_+_20795781 | 0.37 |
ENSDART00000128577
|
ttc34
|
tetratricopeptide repeat domain 34 |
| chr1_-_45920632 | 0.37 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr24_-_21674950 | 0.37 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr18_-_26977940 | 0.37 |
ENSDART00000147823
|
rcn2
|
reticulocalbin 2 |
| chr2_+_16160906 | 0.36 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr23_-_306796 | 0.36 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr13_+_233482 | 0.36 |
ENSDART00000102511
|
cfap36
|
cilia and flagella associated protein 36 |
| chr15_+_3284684 | 0.36 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
Network of associatons between targets according to the STRING database.
First level regulatory network of myf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 1.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.4 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.4 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.6 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.6 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.4 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.4 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.7 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 2.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.1 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 2.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.7 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.1 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.2 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 0.3 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.6 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 1.1 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.2 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.4 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 1.2 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.5 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.0 | 0.6 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.4 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 1.0 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.3 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.3 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.6 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.5 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.5 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 11.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.2 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.1 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.4 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.3 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.2 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.1 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.1 | GO:0016037 | light absorption(GO:0016037) |
| 0.0 | 0.0 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.5 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.3 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.2 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.9 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.1 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.8 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.3 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.0 | 0.0 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.2 | GO:0014020 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 1.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0043198 | ciliary rootlet(GO:0035253) dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 1.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 1.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 0.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.3 | 0.8 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.2 | 1.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 1.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 0.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.5 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 1.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.7 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.7 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.4 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.2 | GO:0042974 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.0 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 9.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |