Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
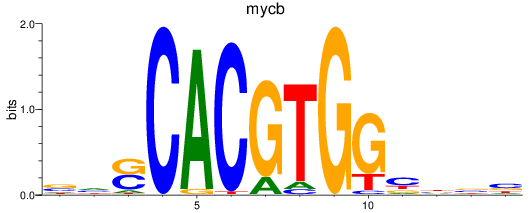
Results for mycb
Z-value: 0.55
Transcription factors associated with mycb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mycb
|
ENSDARG00000007241 | MYC proto-oncogene, bHLH transcription factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mycb | dr11_v1_chr2_+_32016516_32016516 | 0.86 | 5.2e-06 | Click! |
Activity profile of mycb motif
Sorted Z-values of mycb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_26834043 | 1.73 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr15_-_17099560 | 0.99 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr9_-_27391908 | 0.96 |
ENSDART00000135221
|
nepro
|
nucleolus and neural progenitor protein |
| chr8_-_46386024 | 0.93 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr12_-_4301234 | 0.84 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr13_-_35907768 | 0.84 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr9_-_11587070 | 0.81 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr20_-_25631256 | 0.76 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr1_-_19648227 | 0.76 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr5_-_30080332 | 0.72 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr1_+_5485799 | 0.68 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr3_+_23029934 | 0.67 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr17_-_48705993 | 0.66 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr12_+_33975065 | 0.62 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr24_-_39518599 | 0.61 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr17_+_24318753 | 0.61 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr2_-_42375275 | 0.60 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr5_-_3927692 | 0.60 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr22_+_1440702 | 0.59 |
ENSDART00000165677
|
si:dkeyp-53d3.3
|
si:dkeyp-53d3.3 |
| chr18_+_5850837 | 0.55 |
ENSDART00000013150
|
cog8
|
component of oligomeric golgi complex 8 |
| chr24_+_36317544 | 0.55 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr7_-_49800755 | 0.54 |
ENSDART00000180072
|
fjx1
|
four jointed box 1 |
| chr1_-_26676391 | 0.54 |
ENSDART00000152492
|
trmo
|
tRNA methyltransferase O |
| chr1_-_26675969 | 0.53 |
ENSDART00000054184
|
trmo
|
tRNA methyltransferase O |
| chr21_+_22846757 | 0.52 |
ENSDART00000185766
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr12_-_33359654 | 0.51 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_-_12888082 | 0.50 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr17_+_51682429 | 0.49 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr9_+_38158570 | 0.49 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr21_+_4509483 | 0.48 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr14_+_22076596 | 0.48 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr4_-_14192254 | 0.40 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr15_+_6652396 | 0.40 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr11_+_25485774 | 0.39 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr19_+_42983613 | 0.39 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr20_+_25625872 | 0.38 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr9_+_907459 | 0.38 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr12_-_33359052 | 0.38 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_40443118 | 0.38 |
ENSDART00000190675
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr2_+_9946121 | 0.38 |
ENSDART00000100696
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr4_-_2945306 | 0.37 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr8_+_26059677 | 0.37 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr11_+_3959495 | 0.36 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr6_-_39080630 | 0.36 |
ENSDART00000021520
ENSDART00000128308 |
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr22_-_28777557 | 0.35 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr13_-_3516473 | 0.34 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr2_-_20715094 | 0.33 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr9_-_27719998 | 0.33 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr14_-_28568107 | 0.32 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr13_-_280652 | 0.32 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr10_+_29771256 | 0.32 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr22_-_28777374 | 0.31 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr18_+_42970208 | 0.31 |
ENSDART00000084454
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr10_-_14929630 | 0.31 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr10_+_29850330 | 0.31 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr13_+_13930263 | 0.31 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr10_+_29849977 | 0.31 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr13_-_280827 | 0.30 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr24_+_16905188 | 0.29 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr8_+_31799410 | 0.29 |
ENSDART00000141226
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr8_-_13678415 | 0.29 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr18_-_5850683 | 0.29 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr9_-_11676491 | 0.28 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr21_+_17768174 | 0.27 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr7_+_41322407 | 0.26 |
ENSDART00000114076
ENSDART00000139093 |
dph2
|
DPH2 homolog (S. cerevisiae) |
| chr7_+_69449814 | 0.25 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr15_-_43978141 | 0.25 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr10_+_2232023 | 0.24 |
ENSDART00000097695
|
cntnap3
|
contactin associated protein like 3 |
| chr5_-_8765428 | 0.24 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr6_+_12006557 | 0.24 |
ENSDART00000128024
|
wdsub1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr11_+_6456146 | 0.24 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr14_-_12822 | 0.24 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr5_-_54714525 | 0.24 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr22_+_17261801 | 0.24 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr14_+_28432737 | 0.24 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr23_-_38160024 | 0.24 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr9_-_27720612 | 0.23 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr19_+_1005933 | 0.22 |
ENSDART00000191953
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr11_-_21303946 | 0.22 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr8_-_39977026 | 0.22 |
ENSDART00000141707
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr22_+_835728 | 0.21 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr5_+_68807170 | 0.20 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr8_-_2246143 | 0.20 |
ENSDART00000135835
|
si:dkeyp-117b11.3
|
si:dkeyp-117b11.3 |
| chr5_-_54714789 | 0.20 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr18_-_5103931 | 0.19 |
ENSDART00000188091
|
pdcd10a
|
programmed cell death 10a |
| chr21_-_11199366 | 0.19 |
ENSDART00000167666
|
dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr1_-_14533611 | 0.19 |
ENSDART00000180678
|
CR847936.6
|
|
| chr2_+_50999675 | 0.19 |
ENSDART00000158064
ENSDART00000165746 ENSDART00000163917 ENSDART00000172038 ENSDART00000169048 ENSDART00000164775 |
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr19_-_26964348 | 0.19 |
ENSDART00000103955
|
zbtb12.2
|
zinc finger and BTB domain containing 12, tandem duplicate 2 |
| chr9_+_33788389 | 0.19 |
ENSDART00000144623
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr1_-_29061285 | 0.18 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr21_+_26071874 | 0.18 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr24_+_32668675 | 0.17 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr6_-_10752937 | 0.17 |
ENSDART00000135093
|
ola1
|
Obg-like ATPase 1 |
| chr3_+_24595922 | 0.17 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr9_+_12887491 | 0.17 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr25_+_30442389 | 0.17 |
ENSDART00000003346
|
pdcd2l
|
programmed cell death 2-like |
| chr11_+_24800156 | 0.17 |
ENSDART00000131976
|
adipor1a
|
adiponectin receptor 1a |
| chr24_-_42090635 | 0.17 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr9_+_27720428 | 0.16 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr14_+_52408619 | 0.15 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr8_-_40251126 | 0.15 |
ENSDART00000180435
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr2_+_50999477 | 0.15 |
ENSDART00000190111
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr7_+_40148492 | 0.15 |
ENSDART00000110789
|
esyt2b
|
extended synaptotagmin-like protein 2b |
| chr9_-_29985390 | 0.14 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr19_+_791538 | 0.14 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr7_+_49800849 | 0.12 |
ENSDART00000164838
ENSDART00000187888 |
PAMR1 (1 of many)
|
si:ch211-102l7.3 |
| chr16_-_38629208 | 0.12 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr22_+_5118361 | 0.12 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr21_-_14832369 | 0.11 |
ENSDART00000144859
|
pus1
|
pseudouridylate synthase 1 |
| chr11_-_21304452 | 0.11 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr23_+_25135858 | 0.11 |
ENSDART00000103986
|
fam3a
|
family with sequence similarity 3, member A |
| chr25_+_17405201 | 0.10 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr17_+_19499157 | 0.10 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr2_+_19236969 | 0.10 |
ENSDART00000163875
ENSDART00000168644 |
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr1_-_44025066 | 0.09 |
ENSDART00000148932
|
si:ch73-109d9.1
|
si:ch73-109d9.1 |
| chr3_+_28939759 | 0.09 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr10_+_39283985 | 0.09 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr6_-_58975010 | 0.09 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr17_-_29877336 | 0.08 |
ENSDART00000166976
|
esrrb
|
estrogen-related receptor beta |
| chr2_+_33796207 | 0.08 |
ENSDART00000124647
|
kiss1ra
|
KISS1 receptor a |
| chr7_-_39751540 | 0.08 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr8_-_22739757 | 0.08 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr13_+_15702142 | 0.07 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr13_+_39277178 | 0.07 |
ENSDART00000113259
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr6_+_12462079 | 0.07 |
ENSDART00000192029
ENSDART00000065385 |
nr4a2b
|
nuclear receptor subfamily 4, group A, member 2b |
| chr11_+_42478184 | 0.07 |
ENSDART00000089963
|
zgc:110286
|
zgc:110286 |
| chr5_-_26950374 | 0.07 |
ENSDART00000050542
|
htra4
|
HtrA serine peptidase 4 |
| chr8_-_30979494 | 0.07 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr10_+_16036573 | 0.07 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr9_+_29985010 | 0.07 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr25_+_17405458 | 0.07 |
ENSDART00000186711
|
e2f4
|
E2F transcription factor 4 |
| chr15_-_12229874 | 0.06 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr19_-_22346582 | 0.06 |
ENSDART00000045675
ENSDART00000169065 |
slc52a2
zgc:109744
|
solute carrier family 52 (riboflavin transporter), member 2 zgc:109744 |
| chr19_-_24758109 | 0.06 |
ENSDART00000161016
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr24_+_10302955 | 0.06 |
ENSDART00000112455
ENSDART00000066794 ENSDART00000149432 |
otulina
otulina
|
OTU deubiquitinase with linear linkage specificity a OTU deubiquitinase with linear linkage specificity a |
| chr10_+_16036246 | 0.06 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr24_+_33392698 | 0.06 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr5_+_72194444 | 0.06 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr12_+_28995942 | 0.06 |
ENSDART00000076334
|
valopb
|
vertebrate ancient long opsin b |
| chr2_+_19236677 | 0.06 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr3_-_28120092 | 0.05 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr7_+_40148213 | 0.05 |
ENSDART00000173705
|
esyt2b
|
extended synaptotagmin-like protein 2b |
| chr5_+_3927989 | 0.05 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr2_-_5199431 | 0.05 |
ENSDART00000063384
|
phb2a
|
prohibitin 2a |
| chr10_-_7913591 | 0.05 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr14_+_5385855 | 0.05 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr8_-_49705058 | 0.04 |
ENSDART00000083760
|
frmd3
|
FERM domain containing 3 |
| chr13_-_9061944 | 0.04 |
ENSDART00000164186
ENSDART00000102051 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr15_-_2632891 | 0.04 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr3_-_47038481 | 0.04 |
ENSDART00000193260
|
CR392045.1
|
|
| chr21_+_3941758 | 0.04 |
ENSDART00000181345
|
setx
|
senataxin |
| chr25_+_10909850 | 0.04 |
ENSDART00000186021
|
CR339041.3
|
|
| chr20_-_25645150 | 0.04 |
ENSDART00000063137
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr21_-_45382112 | 0.04 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr15_+_17621375 | 0.03 |
ENSDART00000158352
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr7_+_20344032 | 0.03 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr10_-_105100 | 0.03 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr15_-_3940671 | 0.03 |
ENSDART00000189741
|
mindy4b
|
MINDY family member 4B |
| chr5_-_61609448 | 0.03 |
ENSDART00000133426
|
si:dkey-261j4.5
|
si:dkey-261j4.5 |
| chr20_+_25445826 | 0.03 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr13_-_18345854 | 0.03 |
ENSDART00000080107
|
si:dkey-228d14.5
|
si:dkey-228d14.5 |
| chr10_-_15963903 | 0.03 |
ENSDART00000142357
|
si:dkey-3h23.3
|
si:dkey-3h23.3 |
| chr25_+_29474982 | 0.02 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr12_-_34716037 | 0.02 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr2_+_39618951 | 0.02 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr8_+_46386601 | 0.02 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr14_+_16287968 | 0.02 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr15_-_14552101 | 0.02 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr19_-_791016 | 0.02 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr13_+_25199849 | 0.02 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr4_+_18824959 | 0.02 |
ENSDART00000146141
ENSDART00000040424 |
slc26a3.1
|
solute carrier family 26 (anion exchanger), member 3 |
| chr1_-_45042210 | 0.02 |
ENSDART00000073694
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr15_-_14589677 | 0.02 |
ENSDART00000172195
|
coq8b
|
coenzyme Q8B |
| chr11_+_25296366 | 0.02 |
ENSDART00000065949
|
top1l
|
DNA topoisomerase I, like |
| chr3_+_57991074 | 0.01 |
ENSDART00000076077
|
myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr12_+_42436328 | 0.01 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr6_-_54078623 | 0.01 |
ENSDART00000154076
|
hyal1
|
hyaluronoglucosaminidase 1 |
| chr7_+_20017211 | 0.01 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr17_+_51627209 | 0.01 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr14_+_4796168 | 0.01 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr20_+_25645459 | 0.01 |
ENSDART00000143555
ENSDART00000036350 |
aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr1_-_54972170 | 0.01 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr19_-_8940068 | 0.01 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr11_+_45299447 | 0.01 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr19_-_32804535 | 0.00 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr21_+_45510448 | 0.00 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr10_+_29260096 | 0.00 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr20_+_35998073 | 0.00 |
ENSDART00000140457
|
opn5
|
opsin 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mycb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 0.6 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 1.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 0.8 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 0.8 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.6 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.5 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.3 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.6 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 0.7 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.4 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:1904035 | activation of MAPKKK activity(GO:0000185) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.9 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 0.8 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 1.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.6 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.7 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.7 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |