Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
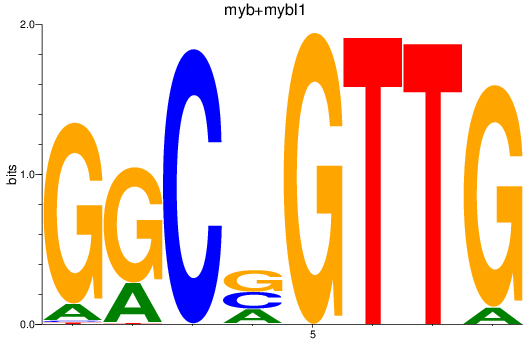
Results for myb+mybl1
Z-value: 1.57
Transcription factors associated with myb+mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mybl1
|
ENSDARG00000030999 | v-myb avian myeloblastosis viral oncogene homolog-like 1 |
|
myb
|
ENSDARG00000053666 | v-myb avian myeloblastosis viral oncogene homolog |
|
myb
|
ENSDARG00000114024 | v-myb avian myeloblastosis viral oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mybl1 | dr11_v1_chr24_+_23791758_23791839 | 0.92 | 9.4e-08 | Click! |
| myb | dr11_v1_chr23_+_31815423_31815492 | 0.69 | 1.6e-03 | Click! |
Activity profile of myb+mybl1 motif
Sorted Z-values of myb+mybl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_21672281 | 3.27 |
ENSDART00000153923
|
si:dkey-40g16.5
|
si:dkey-40g16.5 |
| chr24_-_37568359 | 3.02 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr15_+_21672700 | 2.75 |
ENSDART00000187043
|
si:dkey-40g16.5
|
si:dkey-40g16.5 |
| chr16_-_29557338 | 2.58 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr20_-_53981626 | 2.52 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr17_+_10593398 | 1.99 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr11_+_2642013 | 1.86 |
ENSDART00000111324
|
zgc:193807
|
zgc:193807 |
| chr17_-_33716688 | 1.86 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr11_-_4286010 | 1.84 |
ENSDART00000171558
|
BX248331.1
|
|
| chr20_-_13640598 | 1.80 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr24_-_37003577 | 1.74 |
ENSDART00000137777
|
dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr3_-_31783737 | 1.63 |
ENSDART00000090809
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr20_+_47434709 | 1.56 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr11_-_37995501 | 1.55 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr23_+_39963599 | 1.53 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr1_-_40608392 | 1.53 |
ENSDART00000101671
|
clgn
|
calmegin |
| chr17_-_25649079 | 1.45 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr10_+_35257651 | 1.40 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr3_+_39853788 | 1.40 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr10_+_11038192 | 1.39 |
ENSDART00000047954
|
ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr1_+_22003173 | 1.39 |
ENSDART00000054388
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr8_+_34922981 | 1.36 |
ENSDART00000188445
ENSDART00000188818 ENSDART00000182965 ENSDART00000180718 |
BX005361.1
|
|
| chr18_+_7639401 | 1.36 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr4_+_13568469 | 1.35 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr8_-_50259448 | 1.32 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr1_+_46026457 | 1.26 |
ENSDART00000132705
|
si:ch211-138g9.2
|
si:ch211-138g9.2 |
| chr7_+_22809905 | 1.26 |
ENSDART00000166900
ENSDART00000143455 ENSDART00000126037 |
sf1
|
splicing factor 1 |
| chr3_-_1520657 | 1.22 |
ENSDART00000053201
|
polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr24_-_35707552 | 1.21 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_-_2993095 | 1.20 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr1_-_44704261 | 1.18 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr15_+_36977208 | 1.17 |
ENSDART00000183625
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr1_-_59411901 | 1.15 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr5_+_9112291 | 1.14 |
ENSDART00000135177
|
AL845369.1
|
|
| chr3_-_37082618 | 1.14 |
ENSDART00000026701
ENSDART00000110716 |
tubg1
|
tubulin, gamma 1 |
| chr1_-_45750391 | 1.13 |
ENSDART00000133979
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr6_-_57539141 | 1.12 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr15_+_29393519 | 1.12 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr6_+_120181 | 1.11 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr16_-_13388821 | 1.10 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr25_+_1732838 | 1.09 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr5_+_22633188 | 1.09 |
ENSDART00000128781
|
mtnr1c
|
melatonin receptor 1C |
| chr1_+_27808916 | 1.08 |
ENSDART00000102335
|
pspc1
|
paraspeckle component 1 |
| chr7_+_20109968 | 1.08 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr7_+_24006875 | 1.08 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr5_+_51001985 | 1.06 |
ENSDART00000074638
|
ankrd31
|
ankyrin repeat domain 31 |
| chr16_-_13281380 | 1.05 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr5_+_66220601 | 1.05 |
ENSDART00000129903
|
mfsd10
|
major facilitator superfamily domain containing 10 |
| chr15_-_45246911 | 1.04 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr9_+_38712474 | 1.03 |
ENSDART00000182874
|
kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr13_+_18471546 | 1.03 |
ENSDART00000090712
ENSDART00000127962 |
stox1
|
storkhead box 1 |
| chr7_-_20756013 | 1.01 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr4_+_5318901 | 1.01 |
ENSDART00000067378
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr5_+_31983098 | 1.00 |
ENSDART00000007458
|
ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr11_-_36263886 | 1.00 |
ENSDART00000140397
|
nfya
|
nuclear transcription factor Y, alpha |
| chr16_-_29164379 | 0.98 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr8_+_10823069 | 0.98 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr23_-_32390742 | 0.97 |
ENSDART00000032861
|
ankrd52a
|
ankyrin repeat domain 52a |
| chr11_-_472547 | 0.95 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr12_-_4598237 | 0.94 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chr17_-_10025234 | 0.94 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr12_+_17436904 | 0.94 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr23_+_1349277 | 0.93 |
ENSDART00000173133
ENSDART00000179877 |
utrn
|
utrophin |
| chr5_+_6892195 | 0.93 |
ENSDART00000048201
|
gb:cr929477
|
expressed sequence CR929477 |
| chr20_-_43602650 | 0.91 |
ENSDART00000152939
|
pimr124
|
Pim proto-oncogene, serine/threonine kinase, related 124 |
| chr12_-_628024 | 0.91 |
ENSDART00000158563
ENSDART00000152612 |
wu:fj29h11
si:ch73-301j1.1
|
wu:fj29h11 si:ch73-301j1.1 |
| chr20_+_43602810 | 0.90 |
ENSDART00000142543
|
si:dkey-206f10.1
|
si:dkey-206f10.1 |
| chr4_-_8611841 | 0.90 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr6_-_34958274 | 0.90 |
ENSDART00000113097
|
hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr2_-_14798295 | 0.89 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr11_+_13176568 | 0.89 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr6_-_50685862 | 0.88 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr13_-_27660955 | 0.87 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr17_-_5610514 | 0.87 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr18_-_2668698 | 0.85 |
ENSDART00000157510
|
relt
|
RELT, TNF receptor |
| chr8_-_34767412 | 0.85 |
ENSDART00000164901
|
CABZ01049825.1
|
|
| chr16_-_27566552 | 0.85 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr19_+_10831362 | 0.84 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr11_-_22361306 | 0.84 |
ENSDART00000180688
ENSDART00000182200 ENSDART00000006580 |
tfeb
|
transcription factor EB |
| chr1_-_44701313 | 0.84 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr22_-_38274188 | 0.83 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_+_21817859 | 0.82 |
ENSDART00000143832
ENSDART00000141462 |
ikzf5
|
IKAROS family zinc finger 5 |
| chr5_-_33259079 | 0.81 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr12_+_9880493 | 0.81 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr11_-_4298288 | 0.80 |
ENSDART00000188239
ENSDART00000183705 ENSDART00000187948 |
CT033790.1
|
|
| chr2_+_30969029 | 0.80 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr5_+_27432958 | 0.79 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr17_-_11418513 | 0.79 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr6_+_58280936 | 0.79 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr3_+_24459709 | 0.79 |
ENSDART00000180976
|
cbx6b
|
chromobox homolog 6b |
| chr22_-_25033105 | 0.78 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr18_-_15551360 | 0.78 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr14_-_31715373 | 0.77 |
ENSDART00000127303
ENSDART00000173274 ENSDART00000173435 ENSDART00000172876 ENSDART00000173036 |
map7d3
|
MAP7 domain containing 3 |
| chr9_-_27857265 | 0.77 |
ENSDART00000177513
ENSDART00000143166 ENSDART00000115313 |
si:rp71-45g20.4
|
si:rp71-45g20.4 |
| chr7_+_30202104 | 0.76 |
ENSDART00000173525
|
wdr76
|
WD repeat domain 76 |
| chr17_-_30473374 | 0.74 |
ENSDART00000155021
|
si:ch211-175f11.5
|
si:ch211-175f11.5 |
| chr13_-_37519774 | 0.74 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr21_+_4256291 | 0.72 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr15_-_1022436 | 0.72 |
ENSDART00000156003
|
znf1010
|
zinc finger protein 1010 |
| chr10_-_25860102 | 0.72 |
ENSDART00000080789
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr17_-_11417904 | 0.71 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr21_+_43178831 | 0.70 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr16_-_42151909 | 0.70 |
ENSDART00000160950
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr16_-_42152145 | 0.69 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr13_+_26703922 | 0.69 |
ENSDART00000020946
|
fancl
|
Fanconi anemia, complementation group L |
| chr11_+_8565828 | 0.69 |
ENSDART00000126523
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr11_+_8565323 | 0.68 |
ENSDART00000081607
ENSDART00000141612 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr12_+_11650146 | 0.68 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr17_-_681142 | 0.66 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr5_-_32424402 | 0.66 |
ENSDART00000077549
|
ncs1a
|
neuronal calcium sensor 1a |
| chr6_-_40744720 | 0.66 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr6_+_40832635 | 0.65 |
ENSDART00000011931
|
ruvbl1
|
RuvB-like AAA ATPase 1 |
| chr4_-_39111612 | 0.64 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr17_+_23968214 | 0.64 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr7_+_20110336 | 0.64 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr16_+_33992418 | 0.62 |
ENSDART00000101885
ENSDART00000130540 |
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr23_-_36724575 | 0.62 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr15_+_4988189 | 0.62 |
ENSDART00000142995
ENSDART00000062852 |
spcs2
|
signal peptidase complex subunit 2 |
| chr6_-_9630081 | 0.62 |
ENSDART00000013066
|
ercc3
|
excision repair cross-complementation group 3 |
| chr13_+_48359573 | 0.61 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr24_-_41180149 | 0.61 |
ENSDART00000019975
|
acvr2ba
|
activin A receptor type 2Ba |
| chr7_-_7823662 | 0.61 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr13_+_4871886 | 0.60 |
ENSDART00000132301
|
micu1
|
mitochondrial calcium uptake 1 |
| chr5_+_44064764 | 0.60 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr24_-_37472727 | 0.60 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr1_-_45920632 | 0.60 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr1_+_22174251 | 0.59 |
ENSDART00000137429
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr3_+_46459540 | 0.59 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr15_-_35134918 | 0.58 |
ENSDART00000157196
|
si:ch211-272b8.7
|
si:ch211-272b8.7 |
| chr24_-_21511737 | 0.58 |
ENSDART00000109587
|
rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr1_+_8442071 | 0.57 |
ENSDART00000143547
|
myo15ab
|
myosin XVAb |
| chr16_+_48616220 | 0.57 |
ENSDART00000004751
ENSDART00000130045 |
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr7_+_56253914 | 0.57 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr21_-_26691959 | 0.57 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr14_-_36862745 | 0.57 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr5_-_57169604 | 0.56 |
ENSDART00000136232
|
si:dkey-79c1.1
|
si:dkey-79c1.1 |
| chr6_-_24392426 | 0.56 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr14_-_15155384 | 0.56 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr20_-_18794789 | 0.56 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr11_-_13341483 | 0.56 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr5_-_18962794 | 0.55 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr21_+_13785975 | 0.55 |
ENSDART00000134682
|
slc31a2
|
solute carrier family 31 (copper transporter), member 2 |
| chr7_+_71586485 | 0.55 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr19_+_10661520 | 0.54 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr25_-_554142 | 0.54 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr1_+_19434198 | 0.54 |
ENSDART00000012552
|
clockb
|
clock circadian regulator b |
| chr16_+_32136550 | 0.54 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr4_-_52165969 | 0.54 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr16_+_38159758 | 0.53 |
ENSDART00000058666
ENSDART00000112165 |
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr21_+_25187210 | 0.53 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr20_+_46699021 | 0.52 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr3_-_31715714 | 0.52 |
ENSDART00000051542
|
ccdc47
|
coiled-coil domain containing 47 |
| chr18_+_15132112 | 0.52 |
ENSDART00000099764
|
zgc:153031
|
zgc:153031 |
| chr23_-_15036789 | 0.51 |
ENSDART00000056570
|
phf20b
|
PHD finger protein 20, b |
| chr22_+_10232527 | 0.51 |
ENSDART00000139297
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr19_-_35450661 | 0.51 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr8_-_25569920 | 0.51 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr25_-_518656 | 0.51 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr2_-_24317240 | 0.51 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr19_-_35450857 | 0.50 |
ENSDART00000179357
|
anln
|
anillin, actin binding protein |
| chr6_-_24392909 | 0.50 |
ENSDART00000171042
ENSDART00000168355 |
brdt
|
bromodomain, testis-specific |
| chr6_+_21740672 | 0.50 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr18_-_15532016 | 0.49 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr17_-_37156520 | 0.49 |
ENSDART00000145669
|
dtnbb
|
dystrobrevin, beta b |
| chr1_+_44540293 | 0.48 |
ENSDART00000003497
ENSDART00000179698 |
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr4_+_2655358 | 0.48 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr16_-_51254694 | 0.48 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr21_-_25748913 | 0.47 |
ENSDART00000134217
|
CR388166.2
|
|
| chr6_-_7720332 | 0.47 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr19_-_41991104 | 0.46 |
ENSDART00000087055
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr23_+_27782071 | 0.46 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr22_+_26263290 | 0.46 |
ENSDART00000184840
|
BX649315.1
|
|
| chr22_-_16658996 | 0.46 |
ENSDART00000138429
|
si:dkey-38n4.2
|
si:dkey-38n4.2 |
| chr22_-_10158038 | 0.45 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr17_+_23938283 | 0.45 |
ENSDART00000184391
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr8_+_30671060 | 0.45 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr9_+_56443416 | 0.45 |
ENSDART00000167947
|
CABZ01079480.1
|
|
| chr13_-_23756700 | 0.45 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr22_-_38274995 | 0.45 |
ENSDART00000179786
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr1_-_53684005 | 0.45 |
ENSDART00000108906
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr19_-_47832853 | 0.45 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr11_-_13341051 | 0.43 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr2_+_47945359 | 0.43 |
ENSDART00000098054
|
ftr19
|
finTRIM family, member 19 |
| chr7_+_53754653 | 0.43 |
ENSDART00000163261
ENSDART00000158160 |
neo1a
|
neogenin 1a |
| chr11_-_22303678 | 0.42 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr8_-_52229462 | 0.42 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr16_+_10422836 | 0.42 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr7_+_14211322 | 0.41 |
ENSDART00000180985
ENSDART00000149938 |
fes
|
FES proto-oncogene, tyrosine kinase |
| chr4_-_36791395 | 0.41 |
ENSDART00000162654
|
si:dkeyp-87d1.1
|
si:dkeyp-87d1.1 |
| chr3_+_27027781 | 0.41 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr13_-_44738574 | 0.40 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr1_-_47089818 | 0.39 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr2_-_38261272 | 0.39 |
ENSDART00000143743
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr20_-_45423498 | 0.39 |
ENSDART00000098424
|
trib2
|
tribbles pseudokinase 2 |
| chr17_+_16873417 | 0.38 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr2_-_11662851 | 0.38 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr19_+_2602903 | 0.38 |
ENSDART00000033132
|
fam126a
|
family with sequence similarity 126, member A |
| chr2_-_59145027 | 0.37 |
ENSDART00000128320
|
FO834803.1
|
|
| chr13_-_45063686 | 0.37 |
ENSDART00000130467
ENSDART00000136679 |
chp1
|
calcineurin-like EF-hand protein 1 |
| chr2_-_58085444 | 0.37 |
ENSDART00000166395
|
fcer1gl
|
Fc receptor, IgE, high affinity I, gamma polypeptide like |
Network of associatons between targets according to the STRING database.
First level regulatory network of myb+mybl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.5 | 2.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.5 | 1.4 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.4 | 1.6 | GO:0099623 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.4 | 1.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.4 | 1.1 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.4 | 1.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 0.9 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 1.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 1.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 0.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.2 | 1.0 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.2 | 0.6 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 1.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.0 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.2 | 1.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.7 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 1.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.4 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.9 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.5 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 2.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.6 | GO:0060420 | box C/D snoRNP assembly(GO:0000492) regulation of heart growth(GO:0060420) |
| 0.1 | 0.6 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 1.3 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 1.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.2 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.4 | GO:0043299 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) regulation of mast cell activation involved in immune response(GO:0033006) leukocyte degranulation(GO:0043299) regulation of leukocyte degranulation(GO:0043300) mast cell degranulation(GO:0043303) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 1.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.0 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0007618 | mating(GO:0007618) |
| 0.1 | 0.4 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.5 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.6 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 2.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.6 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.4 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.1 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.2 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 1.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 1.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 1.0 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.6 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.4 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 1.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.0 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.4 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.5 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 4.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 0.6 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 1.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 0.5 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 0.6 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.4 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 2.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 4.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.2 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 1.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |