Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
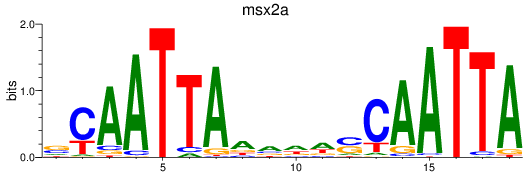
Results for msx2a
Z-value: 0.48
Transcription factors associated with msx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2a
|
ENSDARG00000104651 | muscle segment homeobox 2a |
|
msx2a
|
ENSDARG00000115813 | muscle segment homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2a | dr11_v1_chr14_-_24081929_24081929 | -0.53 | 2.4e-02 | Click! |
Activity profile of msx2a motif
Sorted Z-values of msx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_35541378 | 0.98 |
ENSDART00000183075
ENSDART00000022147 |
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr23_-_44574059 | 0.90 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr16_-_46578523 | 0.89 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr22_+_26853254 | 0.87 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr24_-_38110779 | 0.79 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr2_+_54641644 | 0.78 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr13_+_38226205 | 0.62 |
ENSDART00000144093
ENSDART00000140179 |
si:dkeyp-4c7.3
|
si:dkeyp-4c7.3 |
| chr11_+_3989403 | 0.53 |
ENSDART00000082379
|
spcs1
|
signal peptidase complex subunit 1 |
| chr24_-_20926812 | 0.53 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr6_-_1432200 | 0.52 |
ENSDART00000182901
|
LO018148.1
|
|
| chr3_+_27798094 | 0.50 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr4_-_77260727 | 0.50 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr7_+_59169081 | 0.46 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr15_+_32405959 | 0.44 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr4_+_17844013 | 0.43 |
ENSDART00000019165
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr17_-_11329959 | 0.39 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr11_+_17984167 | 0.39 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr21_+_17005737 | 0.38 |
ENSDART00000101246
|
vps29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr11_+_17984354 | 0.38 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr15_+_32423801 | 0.38 |
ENSDART00000165591
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr8_-_53535262 | 0.37 |
ENSDART00000167839
ENSDART00000157521 |
actr8
|
ARP8 actin related protein 8 homolog |
| chr18_-_17485419 | 0.36 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr5_+_53876169 | 0.36 |
ENSDART00000171107
|
naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr3_-_15496295 | 0.35 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr16_-_55028740 | 0.34 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr16_+_34111919 | 0.34 |
ENSDART00000134037
ENSDART00000006061 ENSDART00000140552 |
tcea3
|
transcription elongation factor A (SII), 3 |
| chr6_+_18359306 | 0.33 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr4_-_14192254 | 0.32 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr20_-_4738101 | 0.32 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr2_-_3611960 | 0.32 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr3_-_15496551 | 0.31 |
ENSDART00000124063
ENSDART00000007726 |
sgf29
|
SAGA complex associated factor 29 |
| chr20_-_26822522 | 0.30 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr1_+_26480890 | 0.29 |
ENSDART00000164430
|
uso1
|
USO1 vesicle transport factor |
| chr8_-_2230128 | 0.29 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr21_+_5192016 | 0.28 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr25_-_35101673 | 0.27 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr5_+_22177033 | 0.27 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr25_-_35101396 | 0.26 |
ENSDART00000138865
|
zgc:162611
|
zgc:162611 |
| chr13_-_17723417 | 0.26 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr17_+_24613255 | 0.26 |
ENSDART00000064738
|
atp5if1b
|
ATP synthase inhibitory factor subunit 1b |
| chr13_+_8693410 | 0.26 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr19_-_5103141 | 0.26 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_-_16152105 | 0.25 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr19_+_31585341 | 0.24 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr10_-_3138403 | 0.24 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr12_-_6063328 | 0.24 |
ENSDART00000002583
|
aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr10_-_35153388 | 0.23 |
ENSDART00000188132
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr22_+_37631234 | 0.23 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr7_+_24528430 | 0.23 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr15_+_3825117 | 0.22 |
ENSDART00000183315
|
CABZ01061591.1
|
|
| chr17_-_31579715 | 0.20 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr15_-_576135 | 0.18 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr7_-_51773166 | 0.18 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr8_+_41048501 | 0.17 |
ENSDART00000123288
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr22_-_27115241 | 0.17 |
ENSDART00000019442
|
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr2_-_54039293 | 0.17 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr15_+_29727799 | 0.16 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr14_-_43616572 | 0.16 |
ENSDART00000111189
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr10_-_42108137 | 0.16 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr20_+_38031439 | 0.15 |
ENSDART00000153208
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr10_-_3138859 | 0.14 |
ENSDART00000190606
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr4_-_6459863 | 0.13 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr9_+_53337974 | 0.13 |
ENSDART00000145138
|
dct
|
dopachrome tautomerase |
| chr6_+_33881372 | 0.13 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr14_+_29941266 | 0.12 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
| chr18_-_46280578 | 0.12 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr16_-_28878080 | 0.12 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr20_+_38032143 | 0.12 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr10_+_20590190 | 0.12 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr6_-_16717878 | 0.12 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr8_+_10561922 | 0.12 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr4_+_5796761 | 0.11 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr23_-_44786844 | 0.11 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr23_-_39784368 | 0.11 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr14_-_498979 | 0.11 |
ENSDART00000171976
|
spry1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr24_+_9881219 | 0.11 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr10_+_20589969 | 0.11 |
ENSDART00000183042
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr17_+_43032529 | 0.11 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr9_-_3934963 | 0.11 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr16_-_36748374 | 0.11 |
ENSDART00000133310
|
pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr14_+_29941445 | 0.11 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr1_-_17569793 | 0.10 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr21_+_29077509 | 0.10 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr20_-_9436521 | 0.09 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr8_-_38616712 | 0.09 |
ENSDART00000141827
|
si:ch211-198d23.1
|
si:ch211-198d23.1 |
| chr16_-_26255877 | 0.09 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr10_+_40781052 | 0.08 |
ENSDART00000191061
ENSDART00000134435 |
taar19c
|
trace amine associated receptor 19c |
| chr25_+_37126921 | 0.08 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr19_+_16016038 | 0.08 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr10_+_40700311 | 0.08 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr10_-_42519773 | 0.07 |
ENSDART00000039187
|
march5l
|
membrane-associated ring finger (C3HC4) 5, like |
| chr7_-_66868543 | 0.07 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr22_+_5532003 | 0.07 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr11_-_165288 | 0.07 |
ENSDART00000108703
ENSDART00000173151 |
tegt
|
testis enhanced gene transcript (BAX inhibitor 1) |
| chr10_+_40756352 | 0.07 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr11_-_3959477 | 0.07 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr22_-_9183944 | 0.07 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr25_+_35019693 | 0.07 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr13_+_18321140 | 0.07 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr12_+_6098713 | 0.07 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr2_-_23677422 | 0.07 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr1_+_8111009 | 0.07 |
ENSDART00000152192
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr10_-_20524387 | 0.06 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr10_-_40454685 | 0.06 |
ENSDART00000137832
|
taar20o
|
trace amine associated receptor 20o |
| chr8_-_16788626 | 0.06 |
ENSDART00000191652
|
CR759968.2
|
|
| chr10_-_40490647 | 0.06 |
ENSDART00000143660
|
taar20x
|
trace amine associated receptor 20x |
| chr10_+_40742685 | 0.06 |
ENSDART00000184858
ENSDART00000140300 |
taar19e
|
trace amine associated receptor 19e |
| chr22_-_24818066 | 0.06 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr21_+_5129513 | 0.06 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr10_-_20524592 | 0.06 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr11_+_42726712 | 0.06 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr10_+_40774215 | 0.06 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr25_+_14507567 | 0.05 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr6_+_8315050 | 0.05 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr7_+_69653981 | 0.05 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr10_+_40670847 | 0.05 |
ENSDART00000109859
|
taar19p
|
trace amine associated receptor 19p |
| chr4_-_25271455 | 0.05 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr7_-_49351706 | 0.05 |
ENSDART00000174151
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr13_+_1369003 | 0.05 |
ENSDART00000146082
|
ifngr1
|
interferon gamma receptor 1 |
| chr1_+_35473397 | 0.05 |
ENSDART00000158799
|
usp38
|
ubiquitin specific peptidase 38 |
| chr10_-_40528229 | 0.05 |
ENSDART00000146927
ENSDART00000148354 |
taar18g
|
trace amine associated receptor 18g |
| chr3_+_48206469 | 0.05 |
ENSDART00000161251
|
si:ch211-207b24.4
|
si:ch211-207b24.4 |
| chr3_+_16771797 | 0.05 |
ENSDART00000147418
|
lrrc3cb
|
leucine rich repeat containing 3Cb |
| chr5_+_36752943 | 0.05 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr10_-_40366360 | 0.05 |
ENSDART00000188830
ENSDART00000192043 ENSDART00000150415 |
taar20i
|
trace amine associated receptor 20i |
| chr1_+_35473219 | 0.05 |
ENSDART00000109678
ENSDART00000181635 |
usp38
|
ubiquitin specific peptidase 38 |
| chr14_-_1987121 | 0.05 |
ENSDART00000135733
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr20_-_38836161 | 0.05 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr6_-_39270851 | 0.05 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr12_+_32199651 | 0.05 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr4_-_23908802 | 0.05 |
ENSDART00000138873
|
celf2
|
cugbp, Elav-like family member 2 |
| chr10_-_40503395 | 0.04 |
ENSDART00000134499
ENSDART00000193101 |
si:dkey-181m19.17
|
si:dkey-181m19.17 |
| chr17_+_23729717 | 0.04 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr19_+_16015881 | 0.04 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr7_-_26408472 | 0.04 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr10_-_40508527 | 0.04 |
ENSDART00000121514
ENSDART00000133729 |
taar20b1
|
trace amine associated receptor 20b1 |
| chr17_+_30448452 | 0.04 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr25_-_18739924 | 0.04 |
ENSDART00000156328
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr5_+_56268436 | 0.04 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr4_+_19700308 | 0.04 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr14_+_45675306 | 0.04 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr10_-_29900546 | 0.04 |
ENSDART00000147441
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr25_+_10909850 | 0.04 |
ENSDART00000186021
|
CR339041.3
|
|
| chr10_+_40665182 | 0.04 |
ENSDART00000099156
|
taar19u
|
trace amine associated receptor 19u |
| chr10_+_40633990 | 0.04 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr19_-_868187 | 0.04 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr7_+_4474880 | 0.04 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr10_+_40762200 | 0.04 |
ENSDART00000161000
ENSDART00000136185 |
taar19g
|
trace amine associated receptor 19g |
| chr10_+_40726399 | 0.04 |
ENSDART00000142812
|
taar19j
|
trace amine associated receptor 19j |
| chr10_-_40360635 | 0.04 |
ENSDART00000131970
|
taar20g
|
trace amine associated receptor 20g |
| chr12_-_25916530 | 0.04 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr21_-_38618540 | 0.04 |
ENSDART00000036600
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr16_-_13921589 | 0.04 |
ENSDART00000023543
|
rcvrn2
|
recoverin 2 |
| chr14_-_1998501 | 0.03 |
ENSDART00000189052
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr10_-_40484531 | 0.03 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr21_-_16113477 | 0.03 |
ENSDART00000147588
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr10_-_17501528 | 0.03 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr4_+_77907740 | 0.03 |
ENSDART00000172216
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr13_+_38990939 | 0.03 |
ENSDART00000145979
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr10_+_40684758 | 0.03 |
ENSDART00000133648
|
taar19h
|
trace amine associated receptor 19h |
| chr7_+_15872357 | 0.03 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr10_+_40568735 | 0.03 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr10_-_40333319 | 0.03 |
ENSDART00000150479
|
taar20a
|
trace amine associated receptor 20a |
| chr10_+_40722256 | 0.03 |
ENSDART00000099154
|
taar19q
|
trace amine associated receptor 19q |
| chr14_+_36869273 | 0.03 |
ENSDART00000111674
|
F2RL3
|
si:ch211-132p1.2 |
| chr10_-_40524741 | 0.03 |
ENSDART00000030565
|
taar18a
|
trace amine associated receptor 18a |
| chr23_-_1660708 | 0.03 |
ENSDART00000175138
|
CU693481.1
|
|
| chr20_-_35557902 | 0.03 |
ENSDART00000134753
|
adgrf7
|
adhesion G protein-coupled receptor F7 |
| chr17_-_36841981 | 0.03 |
ENSDART00000131566
|
myo6b
|
myosin VIb |
| chr14_-_4076480 | 0.03 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr15_+_8767650 | 0.03 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr12_-_29305533 | 0.03 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr10_-_40429747 | 0.03 |
ENSDART00000150613
|
taar20r
|
trace amine associated receptor 20r |
| chr10_-_40406278 | 0.03 |
ENSDART00000131933
ENSDART00000156945 |
taar20i
|
trace amine associated receptor 20i |
| chr20_+_26556372 | 0.03 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr21_-_13751535 | 0.03 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr10_-_34870667 | 0.03 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr7_+_56577522 | 0.02 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr17_+_35362851 | 0.02 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr10_-_40543896 | 0.02 |
ENSDART00000186921
ENSDART00000136031 |
taar18f
|
trace amine associated receptor 18f |
| chr10_-_40402904 | 0.02 |
ENSDART00000145101
|
taar20t
|
trace amine associated receptor 20t |
| chr13_+_40686133 | 0.02 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr5_+_12528693 | 0.02 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr18_+_13837746 | 0.02 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr16_+_28578352 | 0.02 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr10_-_43844537 | 0.02 |
ENSDART00000114208
|
tlr8b
|
toll-like receptor 8b |
| chr7_-_7493758 | 0.02 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr10_+_40660772 | 0.02 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr10_+_40629616 | 0.02 |
ENSDART00000147476
|
CR396590.9
|
|
| chr22_+_30447969 | 0.02 |
ENSDART00000193066
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr15_+_2520319 | 0.02 |
ENSDART00000063329
|
cux1b
|
cut-like homeobox 1b |
| chr24_-_7699356 | 0.02 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr24_+_14713776 | 0.02 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr10_-_40479911 | 0.02 |
ENSDART00000136741
|
taar20d1
|
trace amine associated receptor 20d1 |
| chr7_+_34790768 | 0.02 |
ENSDART00000075116
|
si:dkey-148a17.6
|
si:dkey-148a17.6 |
| chr7_-_47282853 | 0.02 |
ENSDART00000169299
|
si:ch211-186j3.3
|
si:ch211-186j3.3 |
| chr4_-_9557186 | 0.02 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr11_-_2632747 | 0.02 |
ENSDART00000008133
|
si:ch211-160o17.2
|
si:ch211-160o17.2 |
| chr2_+_48073972 | 0.02 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr1_-_17570013 | 0.02 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.1 | GO:2000648 | neuroblast division(GO:0055057) positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.3 | GO:0034650 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.3 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.2 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |