Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
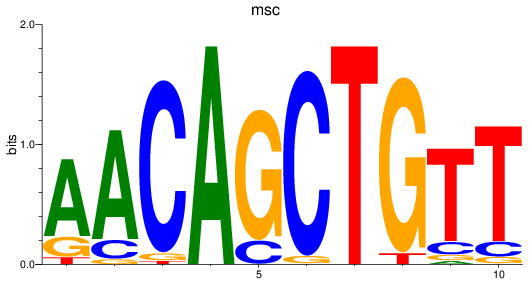
Results for msc
Z-value: 0.48
Transcription factors associated with msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msc
|
ENSDARG00000110016 | musculin (activated B-cell factor-1) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msc | dr11_v1_chr24_+_13735616_13735616 | -0.50 | 3.5e-02 | Click! |
Activity profile of msc motif
Sorted Z-values of msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_11756040 | 0.66 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr3_-_29962345 | 0.62 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr25_-_9013963 | 0.62 |
ENSDART00000073912
|
rag2
|
recombination activating gene 2 |
| chr25_-_19655820 | 0.56 |
ENSDART00000149585
ENSDART00000104353 |
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr3_+_17910569 | 0.54 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr10_-_6867282 | 0.51 |
ENSDART00000144001
ENSDART00000109744 |
ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr16_-_4610255 | 0.47 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr16_-_28727763 | 0.41 |
ENSDART00000149575
|
dcst1
|
DC-STAMP domain containing 1 |
| chr18_+_19820008 | 0.41 |
ENSDART00000079691
ENSDART00000135049 |
iqch
|
IQ motif containing H |
| chr24_-_35707552 | 0.41 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_-_33659328 | 0.40 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr20_-_21672970 | 0.40 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr5_-_15494164 | 0.38 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr1_-_39895859 | 0.38 |
ENSDART00000135791
ENSDART00000035739 |
tmem134
|
transmembrane protein 134 |
| chr6_+_46259950 | 0.36 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr9_-_40873934 | 0.36 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr22_+_35472653 | 0.35 |
ENSDART00000076424
ENSDART00000187204 |
tctex1d2
|
Tctex1 domain containing 2 |
| chr2_-_55779927 | 0.35 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr16_-_13281380 | 0.35 |
ENSDART00000103882
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr9_+_21795917 | 0.35 |
ENSDART00000169069
|
rev1
|
REV1, polymerase (DNA directed) |
| chr7_+_6652967 | 0.34 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr7_+_19552381 | 0.33 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr5_-_22019061 | 0.33 |
ENSDART00000113066
|
amer1
|
APC membrane recruitment protein 1 |
| chr17_+_12075805 | 0.33 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr5_+_25762271 | 0.32 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr10_+_22891126 | 0.32 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr22_+_11775269 | 0.32 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr1_+_27825980 | 0.30 |
ENSDART00000160524
|
pspc1
|
paraspeckle component 1 |
| chr19_+_7636941 | 0.30 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr22_+_17214389 | 0.30 |
ENSDART00000187083
ENSDART00000159468 ENSDART00000129109 ENSDART00000090107 |
nrd1b
|
nardilysin b (N-arginine dibasic convertase) |
| chr7_-_50367642 | 0.30 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr19_-_10214264 | 0.30 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr5_-_29152457 | 0.29 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr5_-_19400166 | 0.29 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr12_-_33646010 | 0.29 |
ENSDART00000111259
|
tmem94
|
transmembrane protein 94 |
| chr4_-_75175407 | 0.29 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr8_-_1219815 | 0.29 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr4_-_20081621 | 0.28 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr7_-_69795488 | 0.28 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr1_-_14258409 | 0.28 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr20_-_33497128 | 0.28 |
ENSDART00000101974
|
erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr1_-_6085750 | 0.27 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr6_+_23931236 | 0.27 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr18_-_14337450 | 0.27 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr16_-_21489514 | 0.26 |
ENSDART00000149525
ENSDART00000148517 ENSDART00000146914 ENSDART00000186493 ENSDART00000193081 ENSDART00000186017 |
mpp6a
|
membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) |
| chr2_+_47471943 | 0.26 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr2_+_47471647 | 0.26 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr4_-_21466480 | 0.26 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr7_-_50367326 | 0.25 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr10_+_26667475 | 0.25 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr7_+_27059330 | 0.25 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr17_+_23968214 | 0.25 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr1_-_24349759 | 0.25 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr20_+_43691208 | 0.25 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr8_-_44299247 | 0.24 |
ENSDART00000144497
|
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr22_+_465269 | 0.24 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr15_-_5624361 | 0.24 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr18_-_14337065 | 0.23 |
ENSDART00000135703
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr11_-_44999858 | 0.23 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr23_-_21758253 | 0.23 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr10_+_31809226 | 0.23 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr19_-_6134802 | 0.23 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr10_-_31805923 | 0.23 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr7_+_17096281 | 0.23 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr14_-_34605804 | 0.22 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr21_+_27382893 | 0.22 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr25_+_36045072 | 0.22 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr8_+_8643901 | 0.22 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr4_-_22335247 | 0.22 |
ENSDART00000192781
|
golgb1
|
golgin B1 |
| chr19_-_23892243 | 0.22 |
ENSDART00000132384
|
si:dkey-222b8.4
|
si:dkey-222b8.4 |
| chr1_+_25801648 | 0.21 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr2_+_1988036 | 0.21 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr9_-_28939181 | 0.21 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr14_+_35464994 | 0.21 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr18_-_7948188 | 0.20 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr8_-_11229523 | 0.20 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr7_-_18470963 | 0.20 |
ENSDART00000173929
ENSDART00000173638 |
znf16l
|
zinc finger protein 16 like |
| chr16_+_28728347 | 0.20 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr16_+_26706519 | 0.19 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr7_-_24364536 | 0.19 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr7_-_59311165 | 0.19 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr7_+_49664174 | 0.19 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr7_+_37742299 | 0.18 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr6_-_35032792 | 0.18 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr22_-_22231720 | 0.18 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr8_+_18615938 | 0.18 |
ENSDART00000089161
|
si:ch211-232d19.4
|
si:ch211-232d19.4 |
| chr16_+_3004422 | 0.17 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr19_+_29808699 | 0.17 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr16_+_40575742 | 0.17 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr17_-_51195651 | 0.17 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr23_-_18057851 | 0.17 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr19_+_29808471 | 0.17 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr1_-_17650223 | 0.16 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr20_-_51831657 | 0.16 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr7_+_67381912 | 0.16 |
ENSDART00000167564
|
nfat5b
|
nuclear factor of activated T cells 5b |
| chr21_+_5974590 | 0.16 |
ENSDART00000098499
|
dck
|
deoxycytidine kinase |
| chr23_-_18057553 | 0.16 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr13_-_31452516 | 0.15 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr14_+_21699129 | 0.15 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr7_+_38962459 | 0.15 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr1_-_55118745 | 0.15 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr20_-_37831849 | 0.15 |
ENSDART00000188483
ENSDART00000153005 ENSDART00000142364 |
si:ch211-147d7.5
|
si:ch211-147d7.5 |
| chr19_-_20446756 | 0.15 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr8_-_14609284 | 0.15 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr20_-_51831816 | 0.14 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr22_-_21897203 | 0.14 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr11_-_35171162 | 0.14 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr15_+_34963316 | 0.14 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr9_+_32306862 | 0.14 |
ENSDART00000078513
|
mob4
|
MOB family member 4, phocein |
| chr10_+_10738880 | 0.14 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr16_-_26676685 | 0.14 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr13_+_7575563 | 0.14 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr18_-_3542435 | 0.14 |
ENSDART00000184017
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr10_-_1697037 | 0.13 |
ENSDART00000125188
ENSDART00000002985 |
srsf9
|
serine/arginine-rich splicing factor 9 |
| chr16_+_28764017 | 0.13 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr14_-_34605607 | 0.13 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr9_+_2020667 | 0.13 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr19_-_32150078 | 0.13 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr15_+_40188076 | 0.13 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr16_+_46401576 | 0.13 |
ENSDART00000130264
|
rpz
|
rapunzel |
| chr1_-_20593778 | 0.13 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr23_+_19790962 | 0.13 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr2_-_51087077 | 0.13 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr14_-_46897067 | 0.12 |
ENSDART00000058789
|
qdpra
|
quinoid dihydropteridine reductase a |
| chr11_+_18873113 | 0.12 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr8_-_44298964 | 0.12 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr2_-_15040345 | 0.12 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr4_+_20255160 | 0.12 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr7_-_41726657 | 0.12 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr15_-_32383529 | 0.12 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr23_-_32162810 | 0.11 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr20_+_23501535 | 0.11 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr6_-_12722360 | 0.11 |
ENSDART00000090174
|
dock9b
|
dedicator of cytokinesis 9b |
| chr20_+_18580176 | 0.11 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr10_-_22803740 | 0.11 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr7_+_20471315 | 0.11 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr22_-_10891213 | 0.11 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr24_-_9300160 | 0.11 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr7_-_29356084 | 0.11 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr17_-_19466319 | 0.11 |
ENSDART00000170429
|
dicer1
|
dicer 1, ribonuclease type III |
| chr4_+_20263097 | 0.11 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr16_-_42750295 | 0.11 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr7_+_38962207 | 0.11 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr4_-_18635005 | 0.11 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr16_+_11151699 | 0.11 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr5_-_13315726 | 0.11 |
ENSDART00000143364
|
sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr14_+_30730749 | 0.11 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr13_-_31164749 | 0.11 |
ENSDART00000049180
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr25_-_25619550 | 0.10 |
ENSDART00000150400
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr7_+_10610791 | 0.10 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr12_-_31103187 | 0.10 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr2_+_37480669 | 0.10 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr18_-_5781922 | 0.10 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr24_+_33802528 | 0.10 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr2_-_7185460 | 0.10 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr20_+_20902549 | 0.10 |
ENSDART00000181195
|
brf1b
|
BRF1, RNA polymerase III transcription initiation factor b |
| chr14_+_38656776 | 0.10 |
ENSDART00000186197
ENSDART00000185861 ENSDART00000020625 |
fbxo38
|
F-box protein 38 |
| chr23_-_27647769 | 0.10 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr23_+_13528550 | 0.10 |
ENSDART00000099903
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr3_+_24537023 | 0.10 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr25_-_36044583 | 0.10 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr6_+_23057311 | 0.10 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr14_+_21699414 | 0.10 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr2_+_37245382 | 0.10 |
ENSDART00000004626
|
sec62
|
SEC62 homolog, preprotein translocation factor |
| chr5_-_17601759 | 0.09 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr17_-_42842129 | 0.09 |
ENSDART00000009528
|
mocs3
|
molybdenum cofactor synthesis 3 |
| chr7_-_7692992 | 0.09 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr12_-_10505986 | 0.09 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr20_+_25645459 | 0.09 |
ENSDART00000143555
ENSDART00000036350 |
aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr10_+_25355308 | 0.09 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr10_-_24371312 | 0.09 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr8_+_21376290 | 0.09 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr7_+_44715224 | 0.09 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr16_-_14353567 | 0.09 |
ENSDART00000139859
|
itga10
|
integrin, alpha 10 |
| chr23_+_35714574 | 0.09 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr16_-_13623928 | 0.09 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr23_+_23658474 | 0.09 |
ENSDART00000162838
|
agrn
|
agrin |
| chr5_-_30475011 | 0.09 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr20_-_26467307 | 0.09 |
ENSDART00000078072
ENSDART00000158213 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr23_-_19831739 | 0.08 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr23_-_30954738 | 0.08 |
ENSDART00000188996
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr8_+_7033049 | 0.08 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr13_+_1575276 | 0.08 |
ENSDART00000165987
|
DST
|
dystonin |
| chr13_+_24834199 | 0.08 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr3_+_22935183 | 0.08 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr2_-_39015130 | 0.08 |
ENSDART00000044331
|
copb2
|
coatomer protein complex, subunit beta 2 |
| chr6_-_10912424 | 0.08 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr3_-_12227359 | 0.08 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr18_-_26797723 | 0.08 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr13_+_42124566 | 0.08 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr18_+_34861568 | 0.08 |
ENSDART00000192825
|
LO018333.1
|
|
| chr7_-_7692723 | 0.08 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr3_-_45245607 | 0.08 |
ENSDART00000181003
ENSDART00000193586 |
usp31
|
ubiquitin specific peptidase 31 |
| chr1_+_2101541 | 0.08 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr9_-_53666031 | 0.08 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr10_-_25069155 | 0.08 |
ENSDART00000078226
ENSDART00000181941 |
mtnr1bb
|
melatonin receptor 1Bb |
| chr11_+_31730680 | 0.07 |
ENSDART00000145497
|
diaph3
|
diaphanous-related formin 3 |
| chr13_+_40815012 | 0.07 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr18_+_35742838 | 0.07 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr15_+_32727848 | 0.07 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
Network of associatons between targets according to the STRING database.
First level regulatory network of msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.3 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.2 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.3 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.3 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.2 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.0 | 0.2 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 0.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.2 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.3 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.0 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.3 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.5 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.5 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |