Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
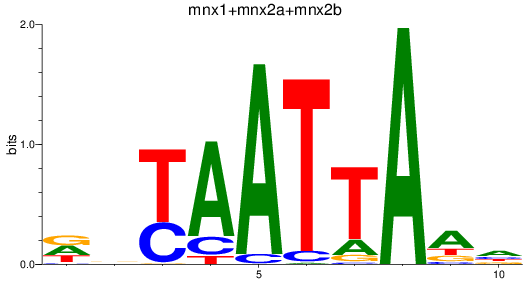
Results for mnx1+mnx2a+mnx2b
Z-value: 0.76
Transcription factors associated with mnx1+mnx2a+mnx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mnx2b
|
ENSDARG00000030350 | motor neuron and pancreas homeobox 2b |
|
mnx1
|
ENSDARG00000035984 | motor neuron and pancreas homeobox 1 |
|
mnx2a
|
ENSDARG00000042106 | motor neuron and pancreas homeobox 2a |
|
mnx1
|
ENSDARG00000109419 | motor neuron and pancreas homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mnx1 | dr11_v1_chr7_+_40638210_40638210 | -0.73 | 6.5e-04 | Click! |
| mnx2a | dr11_v1_chr9_+_7724152_7724152 | -0.56 | 1.7e-02 | Click! |
| mnx2b | dr11_v1_chr1_-_5455498_5455502 | -0.46 | 5.6e-02 | Click! |
Activity profile of mnx1+mnx2a+mnx2b motif
Sorted Z-values of mnx1+mnx2a+mnx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_18130300 | 2.49 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr11_+_18157260 | 2.29 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr10_-_8046764 | 2.16 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr11_-_6452444 | 1.98 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr8_+_45334255 | 1.88 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr10_-_8060573 | 1.74 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr10_-_8053385 | 1.71 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr10_-_21362071 | 1.68 |
ENSDART00000125167
|
avd
|
avidin |
| chr20_-_23426339 | 1.68 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr16_-_29387215 | 1.64 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr10_-_21362320 | 1.58 |
ENSDART00000189789
|
avd
|
avidin |
| chr10_-_25217347 | 1.39 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr11_-_44801968 | 1.11 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr12_+_23912074 | 1.02 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr17_-_40956035 | 1.01 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr16_+_39159752 | 0.86 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr16_-_42056137 | 0.81 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr19_+_42432625 | 0.81 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr3_-_27647845 | 0.81 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr17_+_16046314 | 0.80 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr13_-_8692860 | 0.76 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr22_-_2937503 | 0.76 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr17_+_16090436 | 0.74 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr21_+_1647990 | 0.72 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr14_+_34490445 | 0.67 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr20_-_45060241 | 0.67 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr19_-_44065581 | 0.64 |
ENSDART00000006338
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr13_+_21768447 | 0.60 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr16_+_35916371 | 0.59 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr5_+_44805269 | 0.59 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr10_+_43039947 | 0.58 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr14_-_16151055 | 0.58 |
ENSDART00000162431
|
ptcd3
|
pentatricopeptide repeat domain 3 |
| chr12_+_22580579 | 0.56 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr20_-_37813863 | 0.55 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr3_+_27798094 | 0.55 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr14_+_16151368 | 0.54 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr5_-_25733745 | 0.53 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr6_-_7776612 | 0.52 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr9_+_50001746 | 0.52 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr12_+_45238292 | 0.52 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr25_-_4313699 | 0.51 |
ENSDART00000154038
|
syt7a
|
synaptotagmin VIIa |
| chr24_-_35561672 | 0.51 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr14_+_35405518 | 0.50 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr3_-_16719244 | 0.48 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr19_+_40069524 | 0.48 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr8_+_2487250 | 0.47 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr14_+_16151636 | 0.46 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr1_+_35985813 | 0.43 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr5_+_25733774 | 0.43 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr1_-_54063520 | 0.43 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr17_+_14965570 | 0.43 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr11_-_25257045 | 0.42 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr17_+_28103675 | 0.41 |
ENSDART00000188078
|
zgc:91908
|
zgc:91908 |
| chr13_+_38817871 | 0.41 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr2_+_37227011 | 0.39 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr8_+_25034544 | 0.39 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr4_-_16833518 | 0.39 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr2_-_38363017 | 0.38 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr13_-_25719628 | 0.37 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr20_-_34028967 | 0.36 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr21_-_11654422 | 0.36 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr14_+_35428152 | 0.36 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr16_-_55028740 | 0.35 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr13_-_36034582 | 0.35 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr24_+_21540842 | 0.34 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr2_+_41526904 | 0.33 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr17_+_31221761 | 0.33 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr24_-_30862168 | 0.32 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr22_-_20166660 | 0.32 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr20_-_7128612 | 0.31 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr17_+_30369396 | 0.31 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr6_+_14301214 | 0.30 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr16_+_33902006 | 0.30 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr1_-_32111882 | 0.29 |
ENSDART00000112639
ENSDART00000101958 |
pnpla4
|
patatin-like phospholipase domain containing 4 |
| chr19_+_15441022 | 0.29 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr12_-_13549538 | 0.28 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr17_+_43867889 | 0.27 |
ENSDART00000132673
ENSDART00000167214 |
zgc:66313
|
zgc:66313 |
| chr18_+_15644559 | 0.27 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_-_46777876 | 0.26 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr20_-_16171297 | 0.25 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr2_+_50608099 | 0.24 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr18_+_19456648 | 0.24 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr15_+_31344472 | 0.24 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr10_+_29850330 | 0.24 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr17_-_21162821 | 0.24 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr21_-_14966718 | 0.24 |
ENSDART00000151200
|
mmp17a
|
matrix metallopeptidase 17a |
| chr7_+_69019851 | 0.23 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr10_-_32494499 | 0.23 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr10_+_42521943 | 0.23 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr8_-_25034411 | 0.23 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr5_+_20147830 | 0.22 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr3_+_32365811 | 0.22 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr10_-_32494304 | 0.21 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr14_+_22113331 | 0.21 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr19_+_5480327 | 0.21 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr9_-_50001606 | 0.21 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr18_-_14677936 | 0.20 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr20_+_27712714 | 0.20 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr8_+_2487883 | 0.20 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr22_-_7129631 | 0.19 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr1_-_44812014 | 0.19 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr12_+_32199651 | 0.19 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr15_-_26931541 | 0.19 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr11_-_39118882 | 0.19 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr15_-_17138640 | 0.18 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr22_-_10440688 | 0.18 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr14_-_22113600 | 0.18 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr11_+_33818179 | 0.17 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr22_+_9027884 | 0.17 |
ENSDART00000171839
|
si:ch211-213a13.2
|
si:ch211-213a13.2 |
| chr25_-_20258508 | 0.15 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr1_+_11977426 | 0.15 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr17_-_37395460 | 0.15 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr1_-_9123465 | 0.15 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr3_-_32873641 | 0.13 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr17_-_44249720 | 0.13 |
ENSDART00000156648
|
otx2b
|
orthodenticle homeobox 2b |
| chr10_+_11282883 | 0.13 |
ENSDART00000135355
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr18_-_19456269 | 0.13 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr13_+_38430466 | 0.13 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr20_+_25879826 | 0.12 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr19_+_15440841 | 0.12 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr25_+_13620555 | 0.12 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr4_-_15103646 | 0.12 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr16_+_42471455 | 0.11 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr20_-_43663494 | 0.11 |
ENSDART00000144564
|
BX470188.1
|
|
| chr22_+_16308806 | 0.11 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr2_-_28671139 | 0.11 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr8_-_30979494 | 0.11 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr22_-_36836085 | 0.10 |
ENSDART00000131451
|
cart1
|
cocaine- and amphetamine-regulated transcript 1 |
| chr7_-_23768234 | 0.10 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr6_-_3998199 | 0.10 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr19_+_1835940 | 0.09 |
ENSDART00000167372
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr4_+_3980247 | 0.09 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr15_-_16177603 | 0.09 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr19_-_5103141 | 0.09 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr13_-_31017960 | 0.09 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr15_-_4528326 | 0.08 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr10_-_28028998 | 0.08 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr17_+_23298928 | 0.08 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr18_+_9637744 | 0.08 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr19_+_30884706 | 0.08 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr9_-_14273652 | 0.08 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr12_+_30723778 | 0.08 |
ENSDART00000153291
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr4_-_16334362 | 0.08 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr15_+_6109861 | 0.07 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr1_-_22512063 | 0.07 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr7_+_39418869 | 0.07 |
ENSDART00000169195
|
CT030188.1
|
|
| chr10_-_33297864 | 0.07 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr7_-_44957503 | 0.07 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr24_-_29963858 | 0.07 |
ENSDART00000183442
|
CR352310.1
|
|
| chr20_-_19511700 | 0.07 |
ENSDART00000040191
|
snx17
|
sorting nexin 17 |
| chr19_-_32641725 | 0.07 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr23_+_28809002 | 0.06 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr5_+_32847238 | 0.06 |
ENSDART00000144364
|
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr6_-_51386656 | 0.06 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr23_-_31512496 | 0.06 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr15_+_857148 | 0.06 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr7_-_31321027 | 0.06 |
ENSDART00000186878
|
CR356242.1
|
|
| chr16_+_26777473 | 0.06 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr10_+_33744098 | 0.06 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr15_+_42933236 | 0.06 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr9_+_22485343 | 0.06 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr20_+_31076488 | 0.05 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr23_+_16638639 | 0.05 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr14_+_36521005 | 0.05 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr11_-_25539323 | 0.05 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr25_+_28282274 | 0.05 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr8_-_12867128 | 0.05 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr22_+_30543437 | 0.05 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr21_-_42007213 | 0.05 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr17_+_49500820 | 0.05 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr6_-_40029423 | 0.05 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr22_-_10397600 | 0.04 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr22_-_24818066 | 0.04 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr21_+_28478663 | 0.04 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr8_+_21229718 | 0.04 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr7_+_17229980 | 0.04 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr10_+_40660772 | 0.04 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr19_-_30524952 | 0.04 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr12_+_30367371 | 0.03 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr18_-_33979422 | 0.03 |
ENSDART00000136535
ENSDART00000167698 |
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr8_-_12867434 | 0.03 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr3_-_26806032 | 0.03 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr21_+_5580948 | 0.03 |
ENSDART00000160373
|
ly6m7
|
lymphocyte antigen 6 family member M7 |
| chr9_-_38021889 | 0.03 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr18_+_20560442 | 0.03 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr12_+_5081759 | 0.03 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr11_+_12052791 | 0.02 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr8_-_8446668 | 0.02 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr14_+_25817628 | 0.02 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr6_+_24398907 | 0.02 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr11_-_12998400 | 0.02 |
ENSDART00000018614
|
chrna4b
|
cholinergic receptor, nicotinic, alpha 4b |
| chr4_+_31405646 | 0.02 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr3_+_45368973 | 0.02 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr6_-_11780070 | 0.02 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr10_+_40568735 | 0.01 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr12_+_30367079 | 0.01 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr22_-_392224 | 0.01 |
ENSDART00000124720
|
CABZ01072989.1
|
|
| chr8_-_30204650 | 0.01 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr1_+_33969015 | 0.01 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr23_+_25354856 | 0.01 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mnx1+mnx2a+mnx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.6 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.4 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 1.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.5 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 1.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 1.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 2.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.8 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 2.4 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.3 | GO:0046461 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 1.6 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 1.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.1 | GO:0032096 | adult feeding behavior(GO:0008343) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.3 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 6.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.3 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.6 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.2 | 5.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 2.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 1.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 1.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |