Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
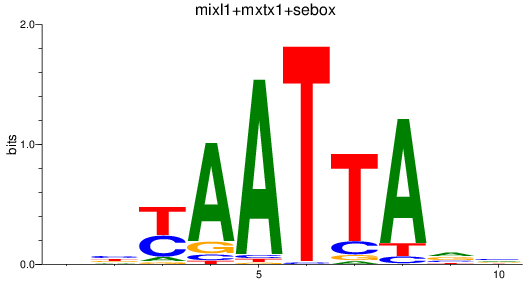
Results for mixl1+mxtx1+sebox
Z-value: 0.56
Transcription factors associated with mixl1+mxtx1+sebox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sebox
|
ENSDARG00000042526 | SEBOX homeobox |
|
mixl1
|
ENSDARG00000069252 | Mix paired-like homeobox |
|
mxtx1
|
ENSDARG00000069382 | mix-type homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mixl1 | dr11_v1_chr20_-_43723860_43723860 | -0.62 | 5.6e-03 | Click! |
| sebox | dr11_v1_chr5_+_67371650_67371650 | -0.33 | 1.8e-01 | Click! |
| mxtx1 | dr11_v1_chr13_-_21660203_21660203 | -0.32 | 2.0e-01 | Click! |
Activity profile of mixl1+mxtx1+sebox motif
Sorted Z-values of mixl1+mxtx1+sebox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_23426339 | 2.13 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr1_-_18811517 | 1.41 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr2_+_6253246 | 1.35 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr18_-_40708537 | 1.01 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr3_+_28860283 | 0.96 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr10_-_21362071 | 0.94 |
ENSDART00000125167
|
avd
|
avidin |
| chr8_+_45334255 | 0.88 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr10_-_21362320 | 0.88 |
ENSDART00000189789
|
avd
|
avidin |
| chr16_-_42056137 | 0.87 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr21_+_25777425 | 0.87 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr22_+_4488454 | 0.77 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr16_-_29387215 | 0.73 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr24_-_37640705 | 0.71 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr1_-_55248496 | 0.71 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr20_-_14114078 | 0.67 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr22_-_2937503 | 0.61 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr20_+_52554352 | 0.60 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr17_+_16046132 | 0.60 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr12_-_14143344 | 0.57 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr10_-_25217347 | 0.56 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr17_+_16046314 | 0.55 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr14_-_33481428 | 0.53 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr5_-_20491198 | 0.47 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr7_-_51773166 | 0.46 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr3_-_26244256 | 0.46 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr4_-_5455506 | 0.46 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr20_-_20931197 | 0.45 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr20_-_20930926 | 0.43 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr17_-_40956035 | 0.43 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr14_+_34492288 | 0.43 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr16_+_29509133 | 0.41 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr11_-_41220794 | 0.40 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr21_+_42226113 | 0.39 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr24_+_40860320 | 0.39 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr25_-_27621268 | 0.38 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr18_+_19456648 | 0.37 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr8_+_25034544 | 0.37 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr19_+_7152966 | 0.37 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr16_-_43344859 | 0.36 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr20_-_37813863 | 0.35 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr8_+_36500308 | 0.35 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr14_+_34490445 | 0.34 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr6_+_50381665 | 0.34 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr23_+_31913292 | 0.34 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr23_+_31912882 | 0.34 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr11_-_1550709 | 0.33 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr20_-_16171297 | 0.33 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr10_+_43039947 | 0.32 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr19_+_40069524 | 0.30 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr16_+_33902006 | 0.29 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr1_+_35985813 | 0.29 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr6_+_21001264 | 0.28 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr21_+_34088377 | 0.28 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr24_+_12835935 | 0.28 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr21_-_11654422 | 0.27 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr17_+_37227936 | 0.27 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr2_+_50608099 | 0.27 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr23_-_25135046 | 0.27 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr11_+_33818179 | 0.27 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr21_+_34088110 | 0.27 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr20_-_45062514 | 0.26 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr6_+_40922572 | 0.26 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr5_+_37903790 | 0.25 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr10_-_32494499 | 0.25 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr16_+_47207691 | 0.24 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr13_+_2625150 | 0.24 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr18_-_19456269 | 0.24 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr7_+_28612671 | 0.24 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr3_+_17537352 | 0.24 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr22_-_20166660 | 0.24 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr1_-_43727012 | 0.24 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr25_+_11456696 | 0.24 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr24_+_21540842 | 0.23 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr3_-_16719244 | 0.23 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr10_-_32494304 | 0.23 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr24_-_30862168 | 0.22 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr6_+_50381347 | 0.21 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr20_-_45060241 | 0.21 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr24_+_16547035 | 0.21 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr8_-_37249813 | 0.20 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr19_-_5103141 | 0.20 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr21_-_39177564 | 0.18 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr3_+_32365811 | 0.18 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr20_-_52902693 | 0.17 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr20_-_52338782 | 0.17 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr12_+_20352400 | 0.16 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr20_-_7583486 | 0.16 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr13_+_4225173 | 0.16 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr15_-_18115540 | 0.16 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr22_+_31207799 | 0.15 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr25_+_31276842 | 0.15 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr18_-_29896367 | 0.15 |
ENSDART00000191303
|
cmc2
|
C-x(9)-C motif containing 2 |
| chr6_-_12172424 | 0.15 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr3_-_31079186 | 0.15 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr23_+_39695827 | 0.14 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr21_-_23331619 | 0.13 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr24_-_24724233 | 0.13 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr24_+_32668675 | 0.13 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr2_-_3678029 | 0.13 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr19_-_32710922 | 0.13 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr17_+_43032529 | 0.13 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr21_-_32060993 | 0.13 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr14_-_22113600 | 0.13 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr8_-_37249991 | 0.12 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr15_-_16177603 | 0.12 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr21_+_15592426 | 0.12 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr5_+_52167986 | 0.12 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr22_-_10586191 | 0.12 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr12_-_30359498 | 0.11 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr5_+_36611128 | 0.10 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr19_+_15441022 | 0.10 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr1_-_26444075 | 0.10 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr3_-_29899172 | 0.09 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr16_+_42471455 | 0.09 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr13_+_1132261 | 0.09 |
ENSDART00000146049
|
wdr92
|
WD repeat domain 92 |
| chr15_-_2519640 | 0.09 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr9_-_31278048 | 0.09 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr13_-_24311628 | 0.08 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr11_+_26604224 | 0.08 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr5_-_37871526 | 0.08 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr9_-_746317 | 0.07 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr17_-_16422654 | 0.07 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr17_-_31639845 | 0.07 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr17_+_8799451 | 0.06 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr8_+_11425048 | 0.06 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr21_-_25801956 | 0.06 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr6_-_3982783 | 0.06 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr1_+_135903 | 0.05 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr14_+_22113331 | 0.05 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr4_+_3980247 | 0.05 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr2_+_41526904 | 0.05 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr23_-_17003533 | 0.05 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr16_+_5774977 | 0.04 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr21_-_5205617 | 0.04 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr3_-_50443607 | 0.04 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr8_+_37527575 | 0.04 |
ENSDART00000147239
|
or135-1
|
odorant receptor, family H, subfamily 135, member 1 |
| chr11_+_12052791 | 0.04 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr3_+_18398876 | 0.04 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr13_-_12602920 | 0.04 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr17_+_23298928 | 0.04 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr20_+_19238382 | 0.04 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr15_+_31344472 | 0.04 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr22_+_19552987 | 0.04 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr22_-_19552796 | 0.04 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr9_-_19161982 | 0.03 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr1_+_21731382 | 0.03 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr14_+_36521005 | 0.03 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr22_-_14739491 | 0.03 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr12_+_10631266 | 0.03 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr14_+_45406299 | 0.03 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr1_+_21937201 | 0.03 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr4_-_71912457 | 0.03 |
ENSDART00000179685
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr23_+_26003187 | 0.02 |
ENSDART00000147271
|
wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr18_+_20560442 | 0.02 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr21_+_44857293 | 0.02 |
ENSDART00000134365
ENSDART00000168217 ENSDART00000065083 |
fstl4
|
follistatin-like 4 |
| chr13_+_38430466 | 0.02 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr1_+_56873359 | 0.02 |
ENSDART00000152713
|
si:ch211-152f2.2
|
si:ch211-152f2.2 |
| chr7_+_56098590 | 0.02 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr13_-_31017960 | 0.02 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr11_+_29770966 | 0.02 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr19_-_30524952 | 0.02 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr15_+_857148 | 0.02 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr10_+_40660772 | 0.02 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr4_+_31405646 | 0.02 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr5_-_50640171 | 0.01 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr5_-_14326959 | 0.01 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr1_+_8694196 | 0.01 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr25_+_3549584 | 0.01 |
ENSDART00000165913
|
ccdc77
|
coiled-coil domain containing 77 |
| chr19_+_30884706 | 0.01 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr6_-_54826061 | 0.01 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr18_+_507835 | 0.01 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr2_+_20332044 | 0.01 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr16_-_29458806 | 0.01 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr8_-_25033681 | 0.01 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr4_-_49582108 | 0.00 |
ENSDART00000154999
|
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr19_-_20113696 | 0.00 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
Network of associatons between targets according to the STRING database.
First level regulatory network of mixl1+mxtx1+sebox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.5 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.2 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 2.3 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.1 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 0.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 2.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.5 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |