Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
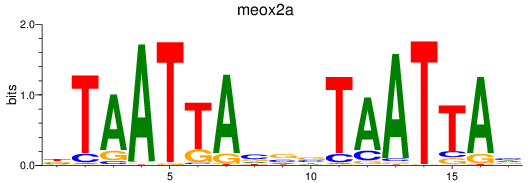
Results for meox2a
Z-value: 0.40
Transcription factors associated with meox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2a
|
ENSDARG00000040911 | mesenchyme homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox2a | dr11_v1_chr15_-_34468599_34468599 | -0.16 | 5.3e-01 | Click! |
Activity profile of meox2a motif
Sorted Z-values of meox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_8060573 | 0.73 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr10_-_8053753 | 0.70 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr13_+_9548849 | 0.68 |
ENSDART00000122695
|
pomk
|
protein-O-mannose kinase |
| chr20_-_6532462 | 0.66 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr22_-_26834043 | 0.61 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr6_-_9952103 | 0.61 |
ENSDART00000065475
|
zp2l2
|
zona pellucida glycoprotein 2, like 2 |
| chr10_-_8053385 | 0.60 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr25_-_4713461 | 0.58 |
ENSDART00000155302
|
drd4a
|
dopamine receptor D4a |
| chr25_+_2361721 | 0.53 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr10_+_14982977 | 0.52 |
ENSDART00000140869
|
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr6_-_32025225 | 0.50 |
ENSDART00000006417
|
pgm1
|
phosphoglucomutase 1 |
| chr23_-_27571667 | 0.50 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr25_-_8660726 | 0.49 |
ENSDART00000168010
ENSDART00000171709 ENSDART00000163426 |
unc45a
|
unc-45 myosin chaperone A |
| chr12_+_34891529 | 0.48 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr16_+_43344475 | 0.45 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr10_+_44699734 | 0.44 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr16_+_54780544 | 0.44 |
ENSDART00000126646
|
si:zfos-1192g2.3
|
si:zfos-1192g2.3 |
| chr10_+_44700103 | 0.43 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr12_+_23912074 | 0.42 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr11_-_2549140 | 0.42 |
ENSDART00000188005
ENSDART00000056065 |
cnpy2
|
canopy 2 |
| chr21_-_14251306 | 0.41 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr3_-_53091946 | 0.40 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr1_-_40132831 | 0.38 |
ENSDART00000135647
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr16_-_43344859 | 0.38 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr3_+_62196672 | 0.37 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr23_-_45398622 | 0.36 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr7_+_51774502 | 0.36 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr7_-_67248829 | 0.35 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr24_-_11076400 | 0.34 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr3_-_25268751 | 0.34 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr12_+_4686145 | 0.33 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr25_-_3759322 | 0.33 |
ENSDART00000088077
|
zgc:158398
|
zgc:158398 |
| chr25_-_17357165 | 0.31 |
ENSDART00000064573
|
setd6
|
SET domain containing 6 |
| chr22_-_16154771 | 0.30 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr13_-_44852324 | 0.30 |
ENSDART00000136455
|
si:dkeyp-2e4.2
|
si:dkeyp-2e4.2 |
| chr12_-_3721957 | 0.30 |
ENSDART00000152433
|
vps25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr21_+_7131970 | 0.29 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr6_-_59942335 | 0.29 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr23_+_42482137 | 0.28 |
ENSDART00000160199
|
cyp2aa3
|
cytochrome P450, family 2, subfamily AA, polypeptide 3 |
| chr24_+_20536056 | 0.28 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr4_-_2168805 | 0.27 |
ENSDART00000150039
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr19_-_47474725 | 0.26 |
ENSDART00000165688
|
ncdn
|
neurochondrin |
| chr4_-_5913338 | 0.26 |
ENSDART00000183590
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr5_-_30074332 | 0.26 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr9_-_4606463 | 0.26 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr17_+_24718272 | 0.25 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr2_-_51644044 | 0.25 |
ENSDART00000157899
|
dad1
|
defender against cell death 1 |
| chr5_-_9073433 | 0.25 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr21_-_43474012 | 0.25 |
ENSDART00000065104
|
tmem185
|
transmembrane protein 185 |
| chr20_-_23494695 | 0.25 |
ENSDART00000058524
ENSDART00000146203 |
anapc4
|
anaphase promoting complex subunit 4 |
| chr12_-_48477031 | 0.24 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr21_-_5881344 | 0.24 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr2_-_21621878 | 0.24 |
ENSDART00000189540
|
zgc:55781
|
zgc:55781 |
| chr6_+_59914126 | 0.24 |
ENSDART00000082325
ENSDART00000102127 |
aamp
|
angio-associated, migratory cell protein |
| chr18_-_34171280 | 0.24 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_-_1258777 | 0.24 |
ENSDART00000077106
ENSDART00000129065 |
ugt5f1
|
UDP glucuronosyltransferase 5 family, polypeptide F1 |
| chr1_-_999556 | 0.23 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr23_-_33709964 | 0.23 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr7_-_46777876 | 0.23 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr4_-_119689 | 0.23 |
ENSDART00000161055
|
dusp16
|
dual specificity phosphatase 16 |
| chr15_-_4580763 | 0.23 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr1_-_24462544 | 0.22 |
ENSDART00000184421
ENSDART00000184987 ENSDART00000126950 |
sh3d19
|
SH3 domain containing 19 |
| chr15_-_2188332 | 0.22 |
ENSDART00000138941
ENSDART00000009564 |
shox2
|
short stature homeobox 2 |
| chr5_+_55984270 | 0.22 |
ENSDART00000047358
ENSDART00000138191 |
fkbp6
|
FK506 binding protein 6 |
| chr19_-_18313303 | 0.21 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr15_-_11683529 | 0.21 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr1_+_24557414 | 0.21 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr21_-_2333357 | 0.21 |
ENSDART00000158743
|
zgc:113348
|
zgc:113348 |
| chr6_-_55585423 | 0.21 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr7_+_52753067 | 0.21 |
ENSDART00000053814
|
mtfmt
|
mitochondrial methionyl-tRNA formyltransferase |
| chr25_-_26753196 | 0.20 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr9_+_3175201 | 0.20 |
ENSDART00000179022
ENSDART00000007756 ENSDART00000182108 |
dync1i2a
|
dynein, cytoplasmic 1, intermediate chain 2a |
| chr21_+_26071874 | 0.19 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr15_-_44077937 | 0.19 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr19_-_47276297 | 0.19 |
ENSDART00000141437
|
sdc2
|
syndecan 2 |
| chr7_+_25126629 | 0.19 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr18_+_50880096 | 0.19 |
ENSDART00000169782
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr18_-_19005919 | 0.19 |
ENSDART00000129776
|
ints14
|
integrator complex subunit 14 |
| chr9_-_52920305 | 0.19 |
ENSDART00000166906
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr19_-_26923957 | 0.19 |
ENSDART00000182390
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr9_-_51436377 | 0.18 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr9_-_52847881 | 0.18 |
ENSDART00000166407
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr3_-_15734358 | 0.18 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr2_+_5371492 | 0.18 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr13_-_17723417 | 0.18 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr21_+_17880511 | 0.18 |
ENSDART00000080481
|
rxraa
|
retinoid X receptor, alpha a |
| chr2_+_4383061 | 0.17 |
ENSDART00000163986
|
wacb
|
WW domain containing adaptor with coiled-coil b |
| chr3_-_29910547 | 0.16 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr3_-_15734530 | 0.16 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr17_-_4252221 | 0.16 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr6_-_1432200 | 0.16 |
ENSDART00000182901
|
LO018148.1
|
|
| chr19_-_8768564 | 0.16 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr19_+_12406583 | 0.16 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr1_+_51721851 | 0.16 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr13_-_18122333 | 0.16 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr5_+_69950882 | 0.16 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr8_-_14080534 | 0.15 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr17_-_21441464 | 0.15 |
ENSDART00000031490
|
vax1
|
ventral anterior homeobox 1 |
| chr20_-_18382708 | 0.15 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr18_+_5875268 | 0.15 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr5_-_26118855 | 0.14 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr24_+_24831727 | 0.14 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr13_+_4368504 | 0.14 |
ENSDART00000108935
|
sft2d1
|
SFT2 domain containing 1 |
| chr3_-_41795917 | 0.14 |
ENSDART00000182662
|
grifin
|
galectin-related inter-fiber protein |
| chr24_+_14937205 | 0.14 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr24_-_20208474 | 0.14 |
ENSDART00000139329
|
cry-dash
|
cryptochrome DASH |
| chr24_-_31904924 | 0.14 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr19_-_44064842 | 0.14 |
ENSDART00000151541
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr19_+_31873308 | 0.13 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr18_+_924949 | 0.13 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr15_+_34946779 | 0.13 |
ENSDART00000192661
ENSDART00000188800 ENSDART00000156515 |
si:ch73-95l15.5
zgc:55621
|
si:ch73-95l15.5 zgc:55621 |
| chr22_+_5971404 | 0.13 |
ENSDART00000122153
|
BX322587.1
|
|
| chr20_-_9428021 | 0.13 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr9_+_30585552 | 0.13 |
ENSDART00000160590
ENSDART00000189815 ENSDART00000143766 |
tbc1d4
|
TBC1 domain family, member 4 |
| chr19_-_40192249 | 0.13 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr5_+_28497956 | 0.13 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr4_-_11737939 | 0.13 |
ENSDART00000150299
|
podxl
|
podocalyxin-like |
| chr23_-_32100106 | 0.13 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr16_+_20161805 | 0.12 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr11_-_20096018 | 0.12 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr2_+_50608099 | 0.12 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr5_-_8096232 | 0.12 |
ENSDART00000158447
|
nipbla
|
nipped-B homolog a (Drosophila) |
| chr8_-_13972626 | 0.12 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr13_-_30713236 | 0.12 |
ENSDART00000112372
ENSDART00000142221 |
tmem72
|
transmembrane protein 72 |
| chr21_-_25685739 | 0.12 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr6_+_27062512 | 0.12 |
ENSDART00000168277
ENSDART00000149429 |
boka
|
BOK, BCL2 family apoptosis regulator a |
| chr12_-_16435150 | 0.12 |
ENSDART00000086211
|
pcgf5b
|
polycomb group ring finger 5b |
| chr13_+_8696825 | 0.12 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr16_-_29403711 | 0.12 |
ENSDART00000175571
|
tlr18
|
toll-like receptor 18 |
| chr23_-_43453975 | 0.12 |
ENSDART00000134142
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr18_-_5875433 | 0.12 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr18_+_814328 | 0.12 |
ENSDART00000157801
ENSDART00000188492 ENSDART00000184489 |
fam219b
|
family with sequence similarity 219, member B |
| chr8_+_49065348 | 0.12 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr12_+_13652361 | 0.12 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr11_-_1509773 | 0.11 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr7_+_67467702 | 0.11 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr21_-_30714665 | 0.11 |
ENSDART00000128011
|
tnfsf10l3
|
tumor necrosis factor (ligand) superfamily, member 10 like 3 |
| chr8_+_47100863 | 0.11 |
ENSDART00000114811
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr2_-_17392799 | 0.11 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr11_+_1512571 | 0.11 |
ENSDART00000027386
|
acot8
|
acyl-CoA thioesterase 8 |
| chr9_-_34882516 | 0.11 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr16_+_43765993 | 0.11 |
ENSDART00000149154
|
si:dkey-122a22.2
|
si:dkey-122a22.2 |
| chr21_+_19345558 | 0.11 |
ENSDART00000184733
|
hpse
|
heparanase |
| chr6_-_52723901 | 0.11 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr17_+_6452511 | 0.11 |
ENSDART00000064694
|
TATDN3
|
Danio rerio TatD DNase domain containing 3-like (LOC571912), mRNA. |
| chr3_+_62327332 | 0.11 |
ENSDART00000158414
|
BX470259.1
|
|
| chr24_+_21540842 | 0.11 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr21_-_11655010 | 0.10 |
ENSDART00000144370
ENSDART00000139814 ENSDART00000139289 |
cast
|
calpastatin |
| chr20_+_26905158 | 0.10 |
ENSDART00000138249
|
rwdd2b
|
RWD domain containing 2B |
| chr14_-_899170 | 0.10 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr3_+_36284986 | 0.10 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr21_-_11654422 | 0.10 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr19_+_3826782 | 0.10 |
ENSDART00000169222
|
oscp1a
|
organic solute carrier partner 1a |
| chr7_+_35075847 | 0.10 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr13_+_45980163 | 0.10 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr1_-_26063188 | 0.10 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr19_-_7525588 | 0.10 |
ENSDART00000092256
|
mrpl9
|
mitochondrial ribosomal protein L9 |
| chr23_-_45705525 | 0.10 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr24_-_33276139 | 0.10 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr7_+_65261576 | 0.10 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr24_-_41320037 | 0.10 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr6_-_10753504 | 0.10 |
ENSDART00000045299
ENSDART00000165645 |
ola1
|
Obg-like ATPase 1 |
| chr8_-_46321889 | 0.10 |
ENSDART00000075189
ENSDART00000122801 |
mtor
|
mechanistic target of rapamycin kinase |
| chr23_-_15284757 | 0.09 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr5_+_29159777 | 0.09 |
ENSDART00000174702
ENSDART00000037891 |
dpp7
|
dipeptidyl-peptidase 7 |
| chr5_+_7279104 | 0.09 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr8_+_20140321 | 0.09 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr13_-_25548733 | 0.09 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr5_+_20147830 | 0.09 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr7_-_17337233 | 0.09 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr24_+_38671054 | 0.09 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr5_+_29160324 | 0.09 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr6_+_18423402 | 0.08 |
ENSDART00000159747
|
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr11_-_11266882 | 0.08 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr14_+_33413980 | 0.08 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr21_-_5205617 | 0.08 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr17_-_16965809 | 0.08 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr23_-_41965557 | 0.08 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr20_-_26066020 | 0.08 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr10_+_3336772 | 0.08 |
ENSDART00000109148
|
brap
|
BRCA1 associated protein |
| chr10_+_26926654 | 0.08 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr5_-_52277643 | 0.08 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr22_-_36530902 | 0.08 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr23_-_3511630 | 0.08 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr9_-_42418470 | 0.08 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr10_-_2524917 | 0.08 |
ENSDART00000188642
|
CU856539.1
|
|
| chr8_-_20243389 | 0.07 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr19_-_29887629 | 0.07 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr11_-_1935883 | 0.07 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr18_+_7286788 | 0.07 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr11_-_7078392 | 0.07 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr14_-_22113600 | 0.07 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr7_+_69905307 | 0.07 |
ENSDART00000035907
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr7_-_51461649 | 0.07 |
ENSDART00000193947
ENSDART00000174328 |
arhgap36
|
Rho GTPase activating protein 36 |
| chr1_+_21725821 | 0.07 |
ENSDART00000132602
|
pax5
|
paired box 5 |
| chr24_+_9178064 | 0.07 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr20_-_9780917 | 0.07 |
ENSDART00000053838
|
cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr22_-_10459880 | 0.06 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr18_-_399554 | 0.06 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr7_+_65398161 | 0.06 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.4 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.7 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.7 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.5 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.6 | GO:0051967 | spinal cord motor neuron cell fate specification(GO:0021520) negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.4 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0090579 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.0 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.0 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 2.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.4 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.4 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |