Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
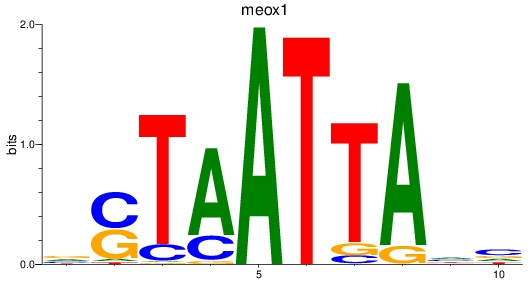
Results for meox1
Z-value: 0.65
Transcription factors associated with meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox1
|
ENSDARG00000007891 | mesenchyme homeobox 1 |
|
meox1
|
ENSDARG00000115382 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox1 | dr11_v1_chr12_+_27462225_27462225 | -0.95 | 2.8e-09 | Click! |
Activity profile of meox1 motif
Sorted Z-values of meox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_40708537 | 2.91 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr6_+_28208973 | 2.62 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr11_-_44801968 | 2.51 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr11_-_1550709 | 2.45 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr23_+_2728095 | 2.42 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr8_-_23780334 | 2.39 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr10_-_34002185 | 2.39 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr2_-_15324837 | 2.30 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr4_-_77125693 | 2.26 |
ENSDART00000174256
|
CU467646.3
|
|
| chr2_-_50225411 | 2.25 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr20_-_6532462 | 2.12 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr10_-_21362071 | 2.07 |
ENSDART00000125167
|
avd
|
avidin |
| chr6_+_21001264 | 2.04 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr4_-_77140030 | 1.98 |
ENSDART00000174332
ENSDART00000190394 ENSDART00000189208 |
CU467646.5
CU467646.7
|
|
| chr9_-_35633827 | 1.98 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr10_-_21545091 | 1.95 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr10_-_21362320 | 1.93 |
ENSDART00000189789
|
avd
|
avidin |
| chr11_-_6452444 | 1.72 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr20_-_23426339 | 1.58 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_16046132 | 1.39 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr12_-_33357655 | 1.36 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr4_+_13586689 | 1.36 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr2_+_15048410 | 1.35 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr14_-_33481428 | 1.32 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr17_+_16046314 | 1.28 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_+_17828267 | 1.27 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr19_-_18127808 | 1.25 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr10_+_16036246 | 1.19 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr11_-_23687158 | 1.17 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr19_-_18127629 | 1.12 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr10_-_35257458 | 1.10 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr20_-_38787047 | 1.02 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr14_+_34490445 | 1.01 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr20_+_29209926 | 0.99 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr2_-_21819421 | 0.98 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr20_-_38787341 | 0.95 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr4_+_13586455 | 0.95 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr2_-_26596794 | 0.94 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr11_-_34219211 | 0.93 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr2_+_6253246 | 0.93 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr10_-_44560165 | 0.91 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr15_-_1038193 | 0.90 |
ENSDART00000159462
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr1_-_23308225 | 0.89 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr16_+_40043673 | 0.88 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr10_+_8197827 | 0.87 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr20_+_29209767 | 0.87 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr7_-_59165640 | 0.86 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr9_-_38021889 | 0.83 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr13_+_38814521 | 0.81 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr22_-_21897203 | 0.80 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr17_+_21902058 | 0.79 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr3_-_26806032 | 0.79 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr2_+_1714640 | 0.76 |
ENSDART00000086761
ENSDART00000111613 |
adgrl2b.1
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr25_-_6049339 | 0.76 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr8_-_15129573 | 0.73 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr20_+_29209615 | 0.73 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr19_+_4968947 | 0.73 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr8_-_30338872 | 0.71 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr20_+_46620374 | 0.71 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr23_-_36446307 | 0.71 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr8_-_25034411 | 0.71 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr15_+_21262917 | 0.71 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr6_-_12172424 | 0.67 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr24_-_34680956 | 0.66 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr12_+_22580579 | 0.65 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr5_-_30074332 | 0.64 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr20_-_20931197 | 0.64 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_-_37744430 | 0.61 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr17_+_49500820 | 0.61 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr10_+_7709724 | 0.60 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr15_+_29140126 | 0.60 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr23_-_10175898 | 0.59 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr24_-_25004553 | 0.58 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr25_+_35891342 | 0.57 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr6_+_4370935 | 0.56 |
ENSDART00000192368
|
rbm26
|
RNA binding motif protein 26 |
| chr15_+_28410664 | 0.56 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr2_-_27619954 | 0.56 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr5_-_68093169 | 0.55 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr7_-_64589920 | 0.53 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr3_-_26805455 | 0.52 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr20_-_20930926 | 0.51 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr13_+_38817871 | 0.50 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr5_-_19006290 | 0.48 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr20_+_13403281 | 0.48 |
ENSDART00000045183
ENSDART00000127350 |
enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr4_-_2380173 | 0.47 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr24_-_41320037 | 0.47 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr12_-_45238759 | 0.47 |
ENSDART00000154859
|
trim65
|
tripartite motif containing 65 |
| chr19_-_19871211 | 0.46 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr17_-_41798856 | 0.45 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr6_-_40713183 | 0.45 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr9_+_43799829 | 0.45 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr20_-_37813863 | 0.45 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr10_+_2582254 | 0.45 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr11_+_31864921 | 0.44 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr17_+_37227936 | 0.44 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr24_+_16547035 | 0.44 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr12_+_48803098 | 0.44 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr12_+_46582727 | 0.44 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr7_+_69459759 | 0.43 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr23_+_33963619 | 0.43 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr12_-_11560794 | 0.43 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr3_-_31254979 | 0.43 |
ENSDART00000130280
|
apnl
|
actinoporin-like protein |
| chr23_-_31913069 | 0.43 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr24_+_39518774 | 0.42 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr7_-_7764287 | 0.42 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr24_+_36204028 | 0.42 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr5_+_40837539 | 0.41 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr14_-_48961056 | 0.41 |
ENSDART00000124192
|
si:dkeyp-121d2.7
|
si:dkeyp-121d2.7 |
| chr19_-_20430892 | 0.41 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr2_-_32825917 | 0.41 |
ENSDART00000180409
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr10_-_41400049 | 0.40 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr4_-_16124417 | 0.40 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr18_-_15551360 | 0.38 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr23_+_7548797 | 0.38 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr12_-_4408828 | 0.38 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr12_-_28349026 | 0.37 |
ENSDART00000183768
ENSDART00000152998 |
zgc:195081
|
zgc:195081 |
| chr6_+_54576520 | 0.37 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr8_-_21142550 | 0.37 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr20_-_28800999 | 0.37 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr17_-_21162821 | 0.36 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr5_-_23117078 | 0.36 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr23_-_900795 | 0.36 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr2_-_32826108 | 0.36 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr22_-_881725 | 0.35 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr14_-_14640401 | 0.35 |
ENSDART00000168027
ENSDART00000167521 |
znf185
|
zinc finger protein 185 with LIM domain |
| chr4_+_25917915 | 0.35 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr2_-_28671139 | 0.35 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr6_-_48418021 | 0.33 |
ENSDART00000090528
|
rhoca
|
ras homolog family member Ca |
| chr3_+_45364849 | 0.33 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr11_+_44135351 | 0.33 |
ENSDART00000182914
|
FO704721.1
|
|
| chr19_+_8612839 | 0.32 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr19_+_12406583 | 0.32 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr1_-_53468160 | 0.32 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr5_+_19314574 | 0.32 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr9_-_5318873 | 0.32 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr15_-_34930727 | 0.31 |
ENSDART00000179723
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr15_-_9272328 | 0.31 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr23_+_4741543 | 0.30 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr20_-_45060241 | 0.30 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr19_-_32641725 | 0.28 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr6_-_2134581 | 0.27 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr18_+_8833251 | 0.26 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr11_+_39107131 | 0.26 |
ENSDART00000105140
|
zgc:112255
|
zgc:112255 |
| chr25_+_10410620 | 0.25 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr11_-_14813029 | 0.25 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr20_-_23226453 | 0.24 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr3_-_31254379 | 0.24 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr18_+_8402129 | 0.23 |
ENSDART00000081154
ENSDART00000171974 |
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr21_-_3853204 | 0.23 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr20_-_9436521 | 0.23 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr25_-_27722309 | 0.23 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr13_+_18321140 | 0.22 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr9_-_3934963 | 0.22 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr19_+_26423590 | 0.22 |
ENSDART00000136244
|
npr1a
|
natriuretic peptide receptor 1a |
| chr17_+_12813316 | 0.21 |
ENSDART00000155629
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr16_-_40043322 | 0.20 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr23_+_42254960 | 0.20 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr9_-_43538328 | 0.20 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr9_+_17983463 | 0.19 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr3_+_27786601 | 0.19 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr17_-_37395460 | 0.19 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr11_+_45323455 | 0.19 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr1_+_51191049 | 0.19 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr12_+_30367371 | 0.19 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr7_-_12464412 | 0.18 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr7_+_21841037 | 0.18 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr8_+_17884569 | 0.18 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr23_-_18130264 | 0.18 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr4_+_9177997 | 0.18 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr22_+_16308450 | 0.18 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr25_-_27722614 | 0.18 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr25_-_4713461 | 0.17 |
ENSDART00000155302
|
drd4a
|
dopamine receptor D4a |
| chr15_-_18432673 | 0.17 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr6_-_55585423 | 0.16 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr6_-_43283122 | 0.15 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr8_+_52314542 | 0.15 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr19_+_15521997 | 0.14 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr24_+_32592748 | 0.14 |
ENSDART00000188256
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr9_+_54695981 | 0.14 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr22_-_3564563 | 0.13 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr11_-_36341028 | 0.13 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr7_-_30174882 | 0.13 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr3_+_45365098 | 0.13 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr15_+_31344472 | 0.13 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr3_-_16719244 | 0.13 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr3_+_25087605 | 0.12 |
ENSDART00000138578
|
st13
|
ST13, Hsp70 interacting protein |
| chr4_-_73587205 | 0.12 |
ENSDART00000150837
|
znf1015
|
zinc finger protein 1015 |
| chr18_+_9637744 | 0.11 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr2_-_39558643 | 0.11 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr5_+_20147830 | 0.11 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr15_+_5280858 | 0.10 |
ENSDART00000174242
|
or121-1
|
odorant receptor, family E, subfamily 121, member 1 |
| chr12_+_30367079 | 0.10 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr20_-_29864390 | 0.10 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr19_+_47476908 | 0.10 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr21_-_3770636 | 0.10 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr8_+_6576940 | 0.10 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr7_+_54642005 | 0.10 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr23_-_31913231 | 0.09 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr21_+_11244068 | 0.09 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr20_+_23185530 | 0.09 |
ENSDART00000142346
|
lrrc66
|
leucine rich repeat containing 66 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.4 | 4.4 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.0 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 2.4 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 0.7 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 0.4 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 2.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.7 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 0.6 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.6 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.5 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 0.4 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 1.0 | GO:0048796 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.4 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.6 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 2.3 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.9 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.8 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.5 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 2.3 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 1.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.9 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.8 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.8 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 1.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 1.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 1.0 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 4.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.7 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 0.9 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.3 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.5 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.4 | GO:0010008 | endosome membrane(GO:0010008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.4 | 2.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 0.9 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 1.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.6 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.9 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.5 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.6 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 1.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 2.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 3.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |