Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
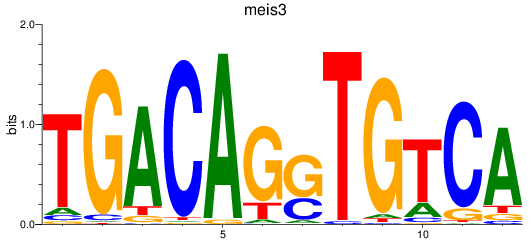
Results for meis3
Z-value: 0.39
Transcription factors associated with meis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis3
|
ENSDARG00000002795 | myeloid ecotropic viral integration site 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis3 | dr11_v1_chr15_-_6863317_6863317 | 0.96 | 2.5e-10 | Click! |
Activity profile of meis3 motif
Sorted Z-values of meis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_25569903 | 1.56 |
ENSDART00000062375
|
si:dkey-48j7.3
|
si:dkey-48j7.3 |
| chr6_+_48041759 | 1.53 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr10_+_26667475 | 1.20 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr20_-_22798794 | 1.09 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr23_+_23658474 | 1.02 |
ENSDART00000162838
|
agrn
|
agrin |
| chr15_-_34056733 | 1.01 |
ENSDART00000170130
ENSDART00000188272 |
VWDE
|
si:dkey-30e9.7 |
| chr2_-_44183613 | 0.98 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr18_+_36769758 | 0.95 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr2_-_44183451 | 0.90 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr3_+_26734162 | 0.87 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr25_-_28600433 | 0.80 |
ENSDART00000138980
|
agbl2
|
ATP/GTP binding protein-like 2 |
| chr25_-_7753207 | 0.78 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr6_+_15762647 | 0.71 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr13_+_22295905 | 0.65 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr8_+_8291492 | 0.64 |
ENSDART00000151314
|
srpk3
|
SRSF protein kinase 3 |
| chr11_+_12879635 | 0.64 |
ENSDART00000182515
ENSDART00000081296 |
si:dkey-11m19.5
|
si:dkey-11m19.5 |
| chr24_+_12989727 | 0.61 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr14_-_26436951 | 0.60 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr20_+_51104367 | 0.60 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr7_+_65673885 | 0.58 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr25_+_32525131 | 0.57 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr18_+_36770166 | 0.57 |
ENSDART00000078151
|
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr5_+_37091626 | 0.57 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr2_+_6127593 | 0.57 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr20_-_23171430 | 0.53 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr21_+_45733871 | 0.45 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr23_+_21663631 | 0.42 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr17_+_35362851 | 0.39 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr20_-_26420939 | 0.38 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr3_-_45308394 | 0.36 |
ENSDART00000155324
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr21_+_30194904 | 0.36 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr25_-_3192405 | 0.36 |
ENSDART00000104835
|
hps5
|
Hermansky-Pudlak syndrome 5 |
| chr20_-_26421112 | 0.35 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr8_-_18225968 | 0.34 |
ENSDART00000135504
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr8_-_7444390 | 0.33 |
ENSDART00000149671
|
hdac6
|
histone deacetylase 6 |
| chr12_+_36413886 | 0.31 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr24_-_26369185 | 0.31 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr11_-_17981421 | 0.31 |
ENSDART00000005999
ENSDART00000104046 |
twf2b
|
twinfilin actin-binding protein 2b |
| chr8_-_7444155 | 0.30 |
ENSDART00000148993
|
hdac6
|
histone deacetylase 6 |
| chr11_-_41966854 | 0.28 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr19_+_34742706 | 0.28 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr6_-_30210378 | 0.27 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr20_-_26467307 | 0.27 |
ENSDART00000078072
ENSDART00000158213 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr22_-_34872533 | 0.27 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr18_-_7810214 | 0.26 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr5_+_24245682 | 0.26 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr11_-_18799827 | 0.26 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr1_-_44084071 | 0.26 |
ENSDART00000166912
|
vwa11
|
von Willebrand factor A domain containing 11 |
| chr7_-_67214972 | 0.25 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr10_-_33379850 | 0.25 |
ENSDART00000186924
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr7_-_26306546 | 0.25 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr6_-_14146979 | 0.24 |
ENSDART00000089564
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr25_-_3139805 | 0.23 |
ENSDART00000166625
|
epx
|
eosinophil peroxidase |
| chr21_+_30351256 | 0.22 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr6_+_3827751 | 0.22 |
ENSDART00000003008
ENSDART00000122348 |
gad1b
|
glutamate decarboxylase 1b |
| chr2_-_32558795 | 0.20 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr8_-_26609259 | 0.19 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr19_-_35596207 | 0.19 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr2_+_9918935 | 0.18 |
ENSDART00000140434
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr23_-_764135 | 0.17 |
ENSDART00000010248
ENSDART00000183978 |
mitfb
|
microphthalmia-associated transcription factor b |
| chr8_-_19280856 | 0.16 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr2_+_9918678 | 0.16 |
ENSDART00000081253
|
acbd5b
|
acyl-CoA binding domain containing 5b |
| chr18_+_660578 | 0.15 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr1_-_40911332 | 0.15 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr6_+_9952678 | 0.14 |
ENSDART00000019325
|
cyp20a1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr11_-_20987378 | 0.14 |
ENSDART00000110140
|
taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr1_+_34496855 | 0.14 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr1_-_10347198 | 0.13 |
ENSDART00000109141
ENSDART00000152565 |
si:ch211-243g6.3
|
si:ch211-243g6.3 |
| chr16_-_41762983 | 0.13 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr5_-_46980651 | 0.13 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr5_+_36752943 | 0.13 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr16_-_26537103 | 0.12 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr23_-_15216654 | 0.11 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr3_+_22443313 | 0.10 |
ENSDART00000156450
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr10_+_406146 | 0.10 |
ENSDART00000145124
|
dact3a
|
dishevelled-binding antagonist of beta-catenin 3a |
| chr8_-_11229523 | 0.09 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr20_+_32406011 | 0.09 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr16_-_1502699 | 0.08 |
ENSDART00000187189
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr13_+_30169681 | 0.07 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr15_-_23442891 | 0.07 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr1_-_26026956 | 0.07 |
ENSDART00000102346
|
si:ch211-145b13.6
|
si:ch211-145b13.6 |
| chr7_+_6879534 | 0.07 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr24_-_30091937 | 0.06 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr15_-_23443588 | 0.05 |
ENSDART00000170427
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr17_-_30839338 | 0.05 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr22_+_30335936 | 0.05 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr16_+_24971717 | 0.05 |
ENSDART00000156958
|
fpr1
|
formyl peptide receptor 1 |
| chr10_-_39011514 | 0.05 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr17_-_43552894 | 0.05 |
ENSDART00000181226
ENSDART00000188125 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr4_+_20812900 | 0.04 |
ENSDART00000005847
|
nav3
|
neuron navigator 3 |
| chr12_-_45876387 | 0.04 |
ENSDART00000043210
ENSDART00000149044 |
pax2b
|
paired box 2b |
| chr2_+_47623202 | 0.04 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr19_+_24872159 | 0.04 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr22_-_5006119 | 0.04 |
ENSDART00000187998
|
rx1
|
retinal homeobox gene 1 |
| chr8_-_8698607 | 0.03 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr8_-_25569920 | 0.03 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr17_+_7595356 | 0.03 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr14_-_9355177 | 0.03 |
ENSDART00000138535
|
fam46d
|
family with sequence similarity 46, member D |
| chr11_-_38513978 | 0.03 |
ENSDART00000123381
|
si:ch211-117k10.3
|
si:ch211-117k10.3 |
| chr17_-_5583345 | 0.02 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr5_-_34964830 | 0.02 |
ENSDART00000133170
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr13_+_45524475 | 0.02 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr2_+_45081489 | 0.02 |
ENSDART00000123966
|
chrng
|
cholinergic receptor, nicotinic, gamma |
| chr5_+_23118470 | 0.02 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr5_+_38913621 | 0.01 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr20_-_26001288 | 0.01 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr19_+_42693855 | 0.01 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr3_+_58656312 | 0.01 |
ENSDART00000148094
|
FP016018.1
|
|
| chr12_+_47122104 | 0.00 |
ENSDART00000184248
|
CABZ01088982.1
|
|
| chr25_-_10610961 | 0.00 |
ENSDART00000153474
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr9_-_25181137 | 0.00 |
ENSDART00000192624
|
esd
|
esterase D/formylglutathione hydrolase |
| chr19_-_30565122 | 0.00 |
ENSDART00000185650
|
hpcal4
|
hippocalcin like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.3 | 1.0 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.2 | 0.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.4 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 1.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.4 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.3 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 1.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.3 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.8 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0019747 | retinoic acid biosynthetic process(GO:0002138) regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |